Kerchunk and Pangeo-Forge
Overview
In this tutorial we are going to use Kerchunk to create reference files of a dataset. This allows us to read an entire dataset as if it were a single Zarr store instead of a collection of NetCDF files. Using Kerchunk, we don’t have to create a copy of the data, instead we create a collection of reference files, so that the original data files can be read as if they were Zarr.
This notebook shares some similarities with the Multi-File Datasets with Kerchunk, as they both create references from NetCDF files. However, this notebook differs as it uses Pangeo-Forge as the runner to create the reference files.
Prerequisites
Concepts |
Importance |
Notes |
|---|---|---|
Kerchunk Basics |
Required |
Core |
Multiple Files and Kerchunk |
Required |
Core |
Kerchunk and Dask |
Required |
Core |
Multi-File Datasets with Kerchunk |
Required |
IO/Visualization |
Time to learn: 45 minutes
Motivation
Why Kerchunk
For many traditional data processing pipelines, the start involves download a large amount of files to a local computer and then subsetting them for future analysis. Kerchunk gives us two large advantages:
A massive reduction in used disk space.
Performance improvements with through parallel, chunk-specific access of the dataset.
In addition to these speedups, once the consolidated Kerchunk reference file has been created, it can be easily shared for other users to access the dataset.
Pangeo-Forge & Kerchunk
Pangeo-Forge is a community project to build reproducible cloud-native ARCO (Analysis-Ready-Cloud-Optimized) datasets. The Python library (pangeo-forge-recipes) is the ETL pipeline to process these datasets or “recipes”. While a majority of the recipes convert a legacy format such as NetCDF to Zarr stores, pangeo-forge-recipes can also use Kerchunk under the hood to create reference recipes.
It is important to note that Kerchunk can be used independently of pangeo-forge-recipes and in this example, pangeo-forge-recipes is acting as the runner for Kerchunk.
Why Pangeo-Forge & Kerchunk
While you can use Kerchunk without pangeo-forge, we hope that pangeo-forge can be another tool to create sharable ARCO datasets using Kerchunk.
A few potential benefits of creating Kerchunk based reference recipes with pangeo-forge may include:
Recipe processing pipelines may be more standardized than case-by-case custom Kerchunk processing functions.
Recipe processing can be scaled through
pangeo-forge-cloudfor large datasets.The infrastructure of
pangeo-forgein GitHub may allow more community feedback on recipes.Additional features such as appending to datasets as new data is generated may be available in future releases of
pangeo-forge.
Getting to Know The Data
gridMET is a high-resolution daily meteorological dataset covering CONUS from 1979-2023. It is produced by the Climatology Lab at UC Merced. In this example, we are going to look create a virtual Zarr dataset of a derived variable, Burn Index.
Create a File Pattern
To build our pangeo-forge pipeline, we need to create a FilePattern object, which is composed of all of our input urls. This dataset ranges from 1979 through 2023 and is composed of one year per file.
To speed up our example, we will prune our recipe to select the first two entries in the FilePattern
from pangeo_forge_recipes.patterns import ConcatDim, FilePattern, MergeDim
years = list(range(1979, 2022 + 1))
time_dim = ConcatDim("time", keys=years)
def format_function(time):
return f"http://www.northwestknowledge.net/metdata/data/bi_{time}.nc"
pattern = FilePattern(format_function, time_dim, file_type="netcdf4")
pattern = pattern.prune()
pattern
<FilePattern {'time': 2}>
Create a Location For Output
We write to local storage for this example, but the reference file could also be shared via cloud storage.
target_root = "references"
store_name = "Pangeo_Forge"
Build the Pangeo-Forge Beam Pipeline
Next, we will chain together a bunch of methods to create a Pangeo-Forge - Apache Beam pipeline.
Processing steps are chained together with the pipe operator (|). Once the pipeline is built, it can be ran in the following cell.
The steps are as follows:
Creates a starting collection of our input file patterns.
Passes those file_patterns to
OpenWithKerchunk, which creates references of each file.Combines the references files into a single reference file with
CombineReferences.Writes the combined reference file.
Note: You can add additional processing steps in this pipeline.
import apache_beam as beam
from pangeo_forge_recipes.transforms import (
OpenWithKerchunk,
WriteCombinedReference,
)
transforms = (
# Create a beam PCollection from our input file pattern
beam.Create(pattern.items())
# Open with Kerchunk and create references for each file
| OpenWithKerchunk(file_type=pattern.file_type)
# Use Kerchunk's `MultiZarrToZarr` functionality to combine and then write references.
# *Note*: Setting the correct contact_dims and identical_dims is important.
| WriteCombinedReference(
target_root=target_root,
store_name=store_name,
concat_dims=["day"],
identical_dims=["lat", "lon", "crs"],
)
)
%%time
with beam.Pipeline() as p:
p | transforms
CPU times: user 1.64 s, sys: 209 ms, total: 1.85 s
Wall time: 20.4 s
import os
import fsspec
full_path = os.path.join(target_root, store_name, "reference.json")
print(os.path.getsize(full_path) / 1e6)
0.048115
Our reference .json file is about 1MB, instead of 108GBs. That is quite the storage savings!
Examine the Result
mapper = fsspec.get_mapper(
"reference://",
fo=full_path,
remote_protocol="http",
)
ds = xr.open_dataset(
mapper, engine="zarr", decode_coords="all", backend_kwargs={"consolidated": False}
)
ds
<xarray.Dataset>
Dimensions: (day: 731, lat: 585, lon: 1386, crs: 1)
Coordinates:
* crs (crs) uint16 3
* day (day) datetime64[ns] 1979-01-01 1979-01-02 ... 1980-12-31
* lat (lat) float64 49.4 49.36 49.32 49.28 ... 25.15 25.11 25.07
* lon (lon) float64 -124.8 -124.7 -124.7 ... -67.14 -67.1 -67.06
Data variables:
burning_index_g (day, lat, lon) float32 ...
Attributes: (12/19)
Conventions: CF-1.6
author: John Abatzoglou - University of Idaho, jabatz...
coordinate_system: EPSG:4326
date: 02 July 2019
geospatial_bounds: POLYGON((-124.7666666333333 49.40000000000000...
geospatial_bounds_crs: EPSG:4326
... ...
geospatial_lon_units: decimal_degrees east
note1: The projection information for this file is: ...
note2: Citation: Abatzoglou, J.T., 2013, Development...
note3: Data in slices after last_permanent_slice (1-...
note4: Data in slices after last_provisional_slice (...
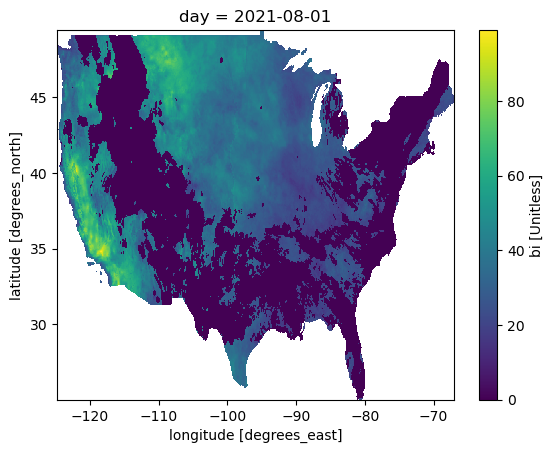
note5: Days correspond approximately to calendar day...ds.isel(day=220).burning_index_g.plot()
<matplotlib.collections.QuadMesh at 0x7fe6509e0b80>
Access Speed Benchmark - Kerchunk vs NetCDF
In the access test below, we had almost a 3x speedup in access time using the Kerchunk reference dataset vs the NetCDF file collection. This isn’t a huge speed-up, but will vary a lot depending on chunking schema, access patterns etc.
Kerchunk |
Time (s) |
|---|---|
Kerchunk |
10 |
Cloud NetCDF |
28 |
Kerchunk
import fsspec
import xarray as xr
%%time
kerchunk_path = os.path.join(target_root, store_name, "reference.json")
mapper = fsspec.get_mapper(
"reference://",
fo=kerchunk_path,
remote_protocol="http",
)
kerchunk_ds = xr.open_dataset(
mapper, engine="zarr", decode_coords="all", backend_kwargs={"consolidated": False}
)
kerchunk_ds.sel(lat=slice(48, 47), lon=slice(-123, -122)).burning_index_g.max().values
CPU times: user 169 ms, sys: 19 ms, total: 188 ms
Wall time: 874 ms
array(61., dtype=float32)
kerchunk_ds
<xarray.Dataset>
Dimensions: (day: 731, lat: 585, lon: 1386, crs: 1)
Coordinates:
* crs (crs) uint16 3
* day (day) datetime64[ns] 1979-01-01 1979-01-02 ... 1980-12-31
* lat (lat) float64 49.4 49.36 49.32 49.28 ... 25.15 25.11 25.07
* lon (lon) float64 -124.8 -124.7 -124.7 ... -67.14 -67.1 -67.06
Data variables:
burning_index_g (day, lat, lon) float32 ...
Attributes: (12/19)
Conventions: CF-1.6
author: John Abatzoglou - University of Idaho, jabatz...
coordinate_system: EPSG:4326
date: 02 July 2019
geospatial_bounds: POLYGON((-124.7666666333333 49.40000000000000...
geospatial_bounds_crs: EPSG:4326
... ...
geospatial_lon_units: decimal_degrees east
note1: The projection information for this file is: ...
note2: Citation: Abatzoglou, J.T., 2013, Development...
note3: Data in slices after last_permanent_slice (1-...
note4: Data in slices after last_provisional_slice (...
note5: Days correspond approximately to calendar day...That took almost 10 seconds.
NetCDF Cloud Access
# prepare urls
def url_gen(year):
return (
f"http://thredds.northwestknowledge.net:8080/thredds/dodsC/MET/bi/bi_{year}.nc"
)
urls_list = [url_gen(year) for year in years]
%%time
netcdf_ds = xr.open_mfdataset(urls_list, engine="netcdf4")
netcdf_ds.sel(lat=slice(48, 47), lon=slice(-123, -122)).burning_index_g.mean().values
CPU times: user 1.17 s, sys: 206 ms, total: 1.37 s
Wall time: 23.5 s
array(11.726682, dtype=float32)
That took about about 28 seconds.
netcdf_ds
<xarray.Dataset>
Dimensions: (lat: 585, lon: 1386, crs: 1, day: 16071)
Coordinates:
* lat (lat) float64 49.4 49.36 49.32 49.28 ... 25.15 25.11 25.07
* lon (lon) float64 -124.8 -124.7 -124.7 ... -67.14 -67.1 -67.06
* crs (crs) float32 3.0
* day (day) datetime64[ns] 1979-01-01 1979-01-02 ... 2022-12-31
Data variables:
burning_index_g (day, lat, lon) float32 dask.array<chunksize=(365, 585, 1386), meta=np.ndarray>
Attributes: (12/19)
geospatial_bounds_crs: EPSG:4326
Conventions: CF-1.6
geospatial_bounds: POLYGON((-124.7666666333333 49.40000000000000...
geospatial_lat_min: 25.066666666666666
geospatial_lat_max: 49.40000000000000
geospatial_lon_min: -124.7666666333333
... ...
date: 02 July 2019
note1: The projection information for this file is: ...
note2: Citation: Abatzoglou, J.T., 2013, Development...
note3: Data in slices after last_permanent_slice (1-...
note4: Data in slices after last_provisional_slice (...
note5: Days correspond approximately to calendar day...5200x Storage Savings
Storage |
Mb (s) |
|---|---|
Kerchunk |
10 |
Cloud NetCDF |
52122 |
# Kerchunk Reference File
import os
print(f"{round(os.path.getsize(kerchunk_path) / 1e6, 1)} Mb")
0.0 Mb
# NetCDF Files
print(f"{round(netcdf_ds.nbytes/1e6,1)} Mb")
print("or")
print(f"{round(netcdf_ds.nbytes/1e9,1)} Gb")
52122.3 Mb
or
52.1 Gb