import uxarray as ux
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import os,sys
import OpenVisus as ov
import openvisuspy as ovp
import xarray as xr
from tqdm import tqdm
import time
from sys import getsizeof
import cmocean
import easygems.healpix as egh
Loading...
# !pip install OpenVisus openvisuspy boto3 xmltodict colorcetidx_filename=f"./idx/conus404_eastward_wind.idx"
def get_size_in_mb(variable):
"""
Calculates the size of a variable in megabytes (MB).
Args:
variable: The variable to check the size of.
Returns:
The size of the variable in MB.
"""
size_in_bytes = sys.getsizeof(variable)
size_in_mb = size_in_bytes / (1024 * 1024)
return size_in_mb
db=ov.LoadDataset(idx_filename)time_to_read_idx=[]
quality=[0,-2,-4,-6,-8,-10,-12,-14,-16]
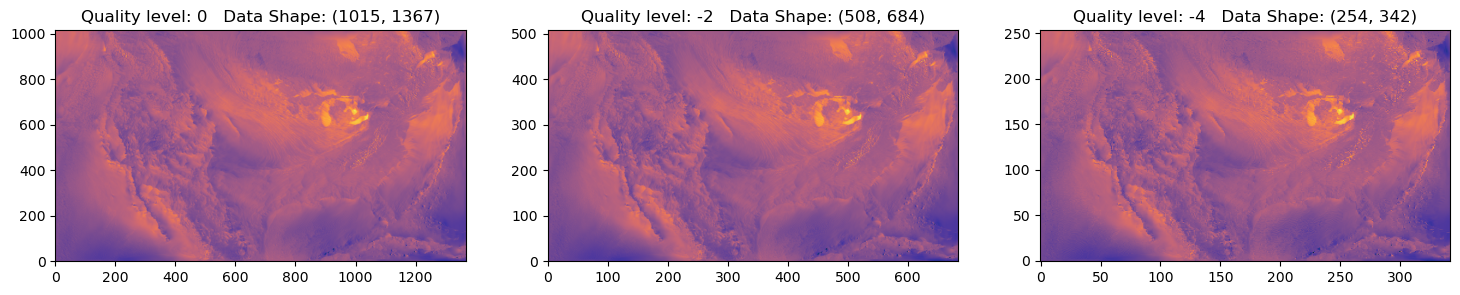
fig, axs = plt.subplots(1, 3, figsize=(18, 3))
for ax,q in zip(axs,quality[0:3]):
start_time=time.time()
data=db.read(quality=q)
end_time=time.time()
total_time=end_time-start_time
time_to_read_idx.append(total_time)
ax.imshow(data,origin='lower',cmap=cmocean.cm.thermal,aspect='auto')
ax.set_title(f'Quality level: {q} Data Shape: {data.shape}')
plt.show()
print(len(time_to_read_idx))
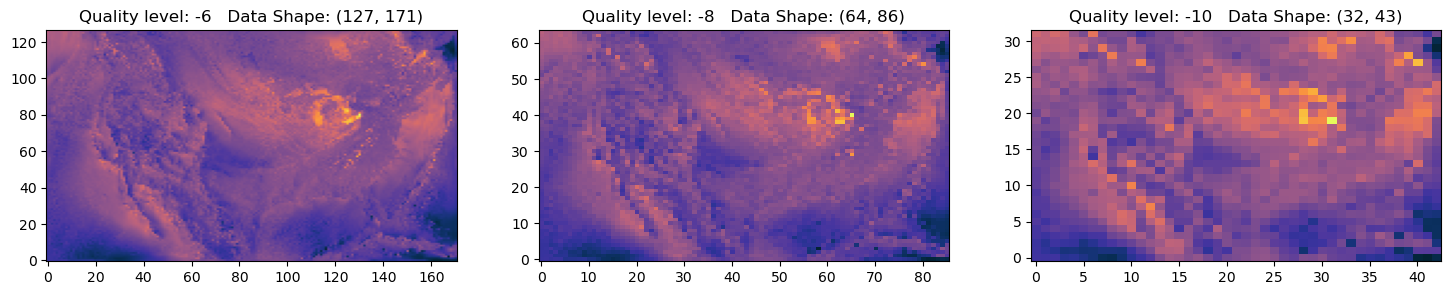
fig, axs = plt.subplots(1, 3, figsize=(18, 3))
for ax,q in zip(axs,quality[3:6]):
start_time=time.time()
data=db.read(quality=q)
end_time=time.time()
total_time=end_time-start_time
time_to_read_idx.append(total_time)
ax.imshow(data,origin='lower',cmap=cmocean.cm.thermal,aspect='auto')
ax.set_title(f'Quality level: {q} Data Shape: {data.shape}')
plt.show()
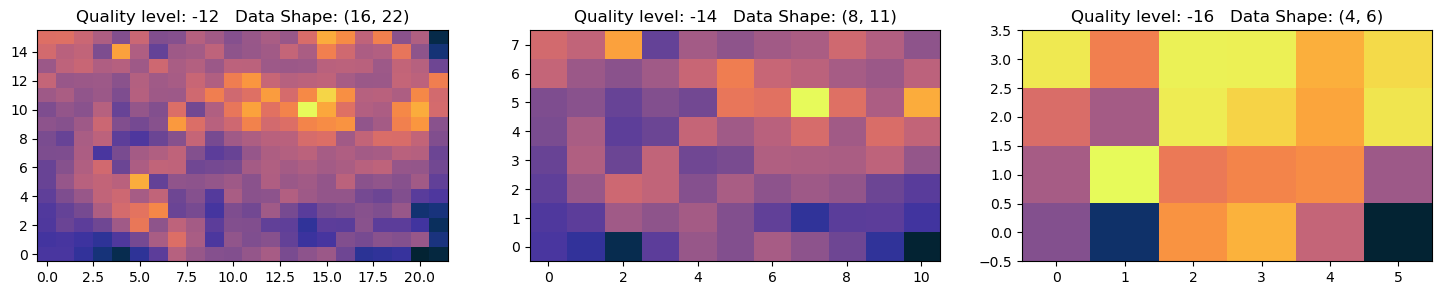
fig, axs = plt.subplots(1, 3, figsize=(18, 3))
for ax,q in zip(axs,quality[6:9]):
start_time=time.time()
data=db.read(quality=q)
end_time=time.time()
total_time=end_time-start_time
time_to_read_idx.append(total_time)
ax.imshow(data,origin='lower',cmap=cmocean.cm.thermal,aspect='auto')
ax.set_title(f'Quality level: {q} Data Shape: {data.shape}')
plt.show()
3
plt.plot(quality,time_to_read_idx)
plt.xlabel('Quality Level')
plt.ylabel('Time in seconds')
plt.title('Time vs quality')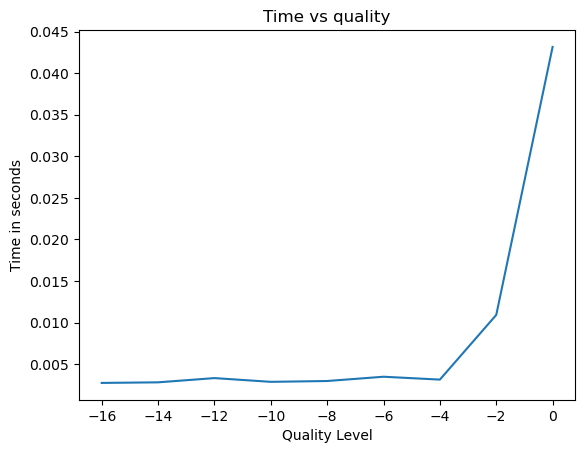
projection = ccrs.Robinson(central_longitude=-100)
fig, ax = plt.subplots(
figsize=(8, 4), subplot_kw={"projection": projection}, constrained_layout=True
)
ax.set_extent([-135, -60, 20, 55], crs=ccrs.PlateCarree())
egh.healpix_show(data, ax=ax, cmap=cmocean.cm.thermal)
ax.add_feature(cf.COASTLINE, linewidth=0.8)
ax.add_feature(cf.BORDERS, linewidth=0.4)---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[7], line 7
2 fig, ax = plt.subplots(
3 figsize=(8, 4), subplot_kw={"projection": projection}, constrained_layout=True
4 )
5 ax.set_extent([-135, -60, 20, 55], crs=ccrs.PlateCarree())
----> 7 egh.healpix_show(data, ax=ax, cmap=cmocean.cm.thermal)
8 ax.add_feature(cf.COASTLINE, linewidth=0.8)
9 ax.add_feature(cf.BORDERS, linewidth=0.4)
File /glade/u/apps/opt/conda/envs/2025-digital-earths-global-hackathon/lib/python3.12/site-packages/easygems/healpix/__init__.py:194, in healpix_show(var, dpi, ax, method, nest, add_coastlines, **kwargs)
192 xlims = ax.get_xlim()
193 ylims = ax.get_ylim()
--> 194 im = healpix_resample(var, xlims, ylims, nx, ny, ax.projection, method, nest)
196 return ax.imshow(im, extent=xlims + ylims, origin="lower", **kwargs)
File /glade/u/apps/opt/conda/envs/2025-digital-earths-global-hackathon/lib/python3.12/site-packages/easygems/healpix/__init__.py:111, in healpix_resample(var, xlims, ylims, nx, ny, src_crs, method, nest)
108 res = np.full(latlon.shape[:-1], np.nan, dtype=var.dtype)
110 if method == "nearest":
--> 111 pix = healpix.ang2pix(
112 get_nside(var),
113 theta=points[0],
114 phi=points[1],
115 nest=nest,
116 lonlat=True,
117 )
118 if var.size < get_npix(var):
119 if not isinstance(var, xr.DataArray):
File /glade/u/apps/opt/conda/envs/2025-digital-earths-global-hackathon/lib/python3.12/site-packages/healpix/__init__.py:69, in ang2pix(nside, theta, phi, nest, lonlat)
68 def ang2pix(nside, theta, phi, nest=False, lonlat=False):
---> 69 check_nside(nside, nest)
70 if not lonlat:
71 if nest:
File /glade/u/apps/opt/conda/envs/2025-digital-earths-global-hackathon/lib/python3.12/site-packages/healpix/__init__.py:29, in check_nside(nside, nest)
27 '''Check whether nside is a valid resolution parameter.'''
28 if nside < 1 or nside > NSIDE_MAX:
---> 29 raise ValueError('nside must be a positive integer 1 <= nside <= '
30 f'2^{np.log2(NSIDE_MAX):.0f}')
31 if nest and (nside & (nside-1)) != 0:
32 raise ValueError('nside must be power of two in the NEST scheme')
ValueError: nside must be a positive integer 1 <= nside <= 2^29