
Overview¶
The first step of any machine learning algorithm is to first load in, filter, and then process the necessary data. Here, we will load in the hurricane track data from IBTRACKS and then filter the tracks by requirements that we will set.
Prerequisites¶
| Concepts | Importance | Notes |
|---|---|---|
| Intro to NUMPY | Necessary | |
| Intro to PANDAS | Necessary | |
| Intro to XARRAY | Necessary | |
| Project management | Helpful |
Time to learn: estimate in minutes. For a rough idea, use 5 mins per subsection, 10 if longer; add these up for a total. Safer to round up and overestimate.
System requirements:
Populate with any system, version, or non-Python software requirements if necessary
Otherwise use the concepts table above and the Imports section below to describe required packages as necessary
If no extra requirements, remove the System requirements point altogether
Imports¶
Begin your body of content with another --- divider before continuing into this section, then remove this body text and populate the following code cell with all necessary Python imports up-front:
import xarray as xr
from dask.distributed import Client
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import pandas as pd
import glob
from global_land_mask import globe
import cartopy.feature as cfeature
from matplotlib.path import Path
import matplotlib.patches as patches
from matplotlib import patheffectsRead in IBTRACKS data¶
First, read in the data as a csv file
ib_data = '../test_folder/ibtracs.NA.list.v04r00.csv'Method to preprocess the track data¶
The following method will set custom ranges for the time period and spatial domain of analysis.
def process_ibrack(ib_loc, periods=[2000,2005]):
#Read in the IBTRACKS data
read_ib_data = pd.read_csv(ib_loc,keep_default_na=False)
#Get the units for each column in read_ib_data
units = read_ib_data.iloc[0,:]
#Get data or the remainder of read_ib_data
ib_original_dft = read_ib_data.iloc[1:,:]
#Set a custom date and time range based on user choosing
ib_original_dft['datetime'] = pd.to_datetime(ib_original_dft['ISO_TIME'],format='%Y-%m-%d %H:%M:%S')
year_mask = (ib_original_dft['datetime'] > f'{periods[0]}-1-1') & (ib_original_dft['datetime'] <= f'{periods[1]}-11-30')
ib_new_period = ib_original_dft[year_mask][ib_original_dft['BASIN'] == 'NA']
#Only use cyclones over the North Atlantic basin
#This can be changed to include more cyclones outside of the Northeast Atlantic
def only_na_basin(df):
lon_wise = df.sort_values(by='datetime')
if lon_wise['LON'].iloc[0] > -55:
return df
else:
return None
#Get the number of time steps in each cyclone, or event
only_neatlantic = ib_new_period.groupby('SID').apply(only_na_basin).reset_index(drop=True)
counts = only_neatlantic.groupby('SID').count().iloc[:,0]
#Get cyclones that last at least 12 time steps
counts_12 = counts[counts > 12].index
persist_storms = ib_new_period[ib_new_period['SID'].isin(counts_12)]
persist_storms['month']= persist_storms['datetime'].dt.month
# mask out land points
def mask_lands(df):
ordered_df = df.sort_values(by='datetime')
lat = ordered_df['LAT']
lon = ordered_df['LON']
ocean_mask = pd.Series(globe.is_ocean(lat=lat,lon=lon))
idx_false = ocean_mask.idxmin()
if idx_false == 0:
return df
else:
land_mask = ocean_mask.iloc[:idx_false]
final_masked = ordered_df.iloc[:idx_false,:]
return final_masked
# filter extratropical parts of the storm tracks
def filter_ET(df):
ordered_df = df.sort_values(by='datetime')
lat_filter = ordered_df['LAT'] <= 35
filter_df = ordered_df[lat_filter]
return filter_df
exclude_et = persist_storms.groupby('SID').apply(filter_ET).reset_index(drop=True)
final_dft = exclude_et.groupby('SID').apply(mask_lands).reset_index(drop=True)
return final_dftProcess IBTRACKS data using the above methods¶
ib_data_processed = process_ibrack(ib_data,periods=[2000,2005])
ib_data_processed['LAT'] = ib_data_processed['LAT'].astype(float)
ib_data_processed['LON'] = ib_data_processed['LON'].astype(float)
ib_data_processed['USA_WIND'] = ib_data_processed['USA_WIND'].astype(float)
ib_data_processed['datetime'] = pd.to_datetime(ib_data_processed['datetime'],format='%Y-%m-%d %H:%M:%S')
ib_data_processed['SID'] = ib_data_processed['SID'].astype(str)/tmp/ipykernel_3888/278484491.py:4: DtypeWarning: Columns (1,2,8,9,14,19,20,23,24,161,162) have mixed types. Specify dtype option on import or set low_memory=False.
read_ib_data = pd.read_csv(ib_loc,keep_default_na=False)
/tmp/ipykernel_3888/278484491.py:13: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
ib_original_dft['datetime'] = pd.to_datetime(ib_original_dft['ISO_TIME'],format='%Y-%m-%d %H:%M:%S')
/tmp/ipykernel_3888/278484491.py:15: UserWarning: Boolean Series key will be reindexed to match DataFrame index.
ib_new_period = ib_original_dft[year_mask][ib_original_dft['BASIN'] == 'NA']
/tmp/ipykernel_3888/278484491.py:27: FutureWarning: DataFrameGroupBy.apply operated on the grouping columns. This behavior is deprecated, and in a future version of pandas the grouping columns will be excluded from the operation. Either pass `include_groups=False` to exclude the groupings or explicitly select the grouping columns after groupby to silence this warning.
only_neatlantic = ib_new_period.groupby('SID').apply(only_na_basin).reset_index(drop=True)
/tmp/ipykernel_3888/278484491.py:33: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
persist_storms['month']= persist_storms['datetime'].dt.month
/tmp/ipykernel_3888/278484491.py:56: FutureWarning: DataFrameGroupBy.apply operated on the grouping columns. This behavior is deprecated, and in a future version of pandas the grouping columns will be excluded from the operation. Either pass `include_groups=False` to exclude the groupings or explicitly select the grouping columns after groupby to silence this warning.
exclude_et = persist_storms.groupby('SID').apply(filter_ET).reset_index(drop=True)
/tmp/ipykernel_3888/278484491.py:58: FutureWarning: DataFrameGroupBy.apply operated on the grouping columns. This behavior is deprecated, and in a future version of pandas the grouping columns will be excluded from the operation. Either pass `include_groups=False` to exclude the groupings or explicitly select the grouping columns after groupby to silence this warning.
final_dft = exclude_et.groupby('SID').apply(mask_lands).reset_index(drop=True)
Organize the edited IBTRACKS file¶
ib_data_processed['id'] = ib_data_processed['SID'].astype('category')
ib_data_processed['id'] = ib_data_processed['id'].cat.codes
req_cols = ['datetime','LAT','LON','USA_WIND','id']
# groupby datetime 6h
ib_data_processed['datetime'] = ib_data_processed['datetime'].dt.floor('6h')
ib_data_processed_6h = ib_data_processed[req_cols].groupby('datetime').mean().reset_index()
# copy the SID based on the id
ib_data_processed_6h['SID'] = ib_data_processed.groupby('datetime')['SID'].first().valuesSee the organized and edited IBTRACKS file¶
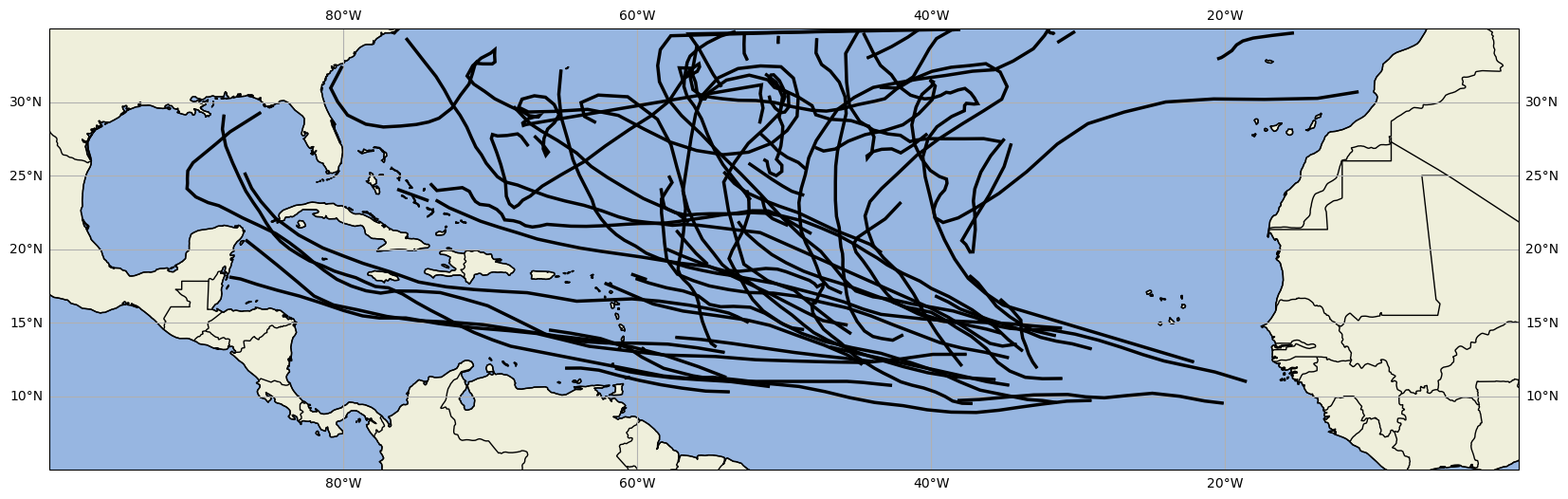
ib_data_processed_6hPlot the track data we have edited and organized¶
def plot_tracks(filtered_ib):
filtered_ib['datetime'] = pd.to_datetime(filtered_ib['datetime'])
events = filtered_ib.groupby('id')
fig,ax = plt.subplots(figsize=(20,10),subplot_kw={"projection": ccrs.PlateCarree()})
ax.add_feature(cfeature.COASTLINE)
ax.add_feature(cfeature.BORDERS)
ax.coastlines()
ax.add_feature(cfeature.LAND)
ax.add_feature(cfeature.OCEAN)
ax.gridlines(draw_labels=True, dms=True, x_inline=False, y_inline=False)
for event_num , event in events:
lon = event['LON'].values
lat = event['LAT'].values
vertices = [(lo, la) for lo, la in zip(lon, lat)]
codes = [Path.MOVETO]
[codes.append(Path.LINETO) for index in range(0, len(event) - 1)]
path = Path(vertices, codes)
patch = patches.PathPatch(path, lw = 1, fc = 'none', path_effects = [patheffects.withStroke(linewidth=2.5, foreground="black")], zorder = 5)
ax.add_patch(patch)
ax.set_xlim(-100, 0)
ax.set_ylim(5, 35)
return fig,ax
plot_tracks(ib_data_processed_6h)(<Figure size 2000x1000 with 1 Axes>, <GeoAxes: >)/home/runner/micromamba/envs/cookbook-dev/lib/python3.12/site-packages/cartopy/io/__init__.py:242: DownloadWarning: Downloading: https://naturalearth.s3.amazonaws.com/50m_physical/ne_50m_land.zip
warnings.warn(f'Downloading: {url}', DownloadWarning)
/home/runner/micromamba/envs/cookbook-dev/lib/python3.12/site-packages/cartopy/io/__init__.py:242: DownloadWarning: Downloading: https://naturalearth.s3.amazonaws.com/50m_physical/ne_50m_ocean.zip
warnings.warn(f'Downloading: {url}', DownloadWarning)
/home/runner/micromamba/envs/cookbook-dev/lib/python3.12/site-packages/cartopy/io/__init__.py:242: DownloadWarning: Downloading: https://naturalearth.s3.amazonaws.com/50m_physical/ne_50m_coastline.zip
warnings.warn(f'Downloading: {url}', DownloadWarning)
/home/runner/micromamba/envs/cookbook-dev/lib/python3.12/site-packages/cartopy/io/__init__.py:242: DownloadWarning: Downloading: https://naturalearth.s3.amazonaws.com/50m_cultural/ne_50m_admin_0_boundary_lines_land.zip
warnings.warn(f'Downloading: {url}', DownloadWarning)
ib_data_processed_6h.to_csv('../test_folder/ib_data_processed_6h.csv')Summary¶
Here, we appropriately edited the cyclone track data over our space and time domain of interest. We have also created an output file that will be useful for eventual creation and training of our machine learning model.
What’s next?¶
Next we will edit the ERA5 data. In particular, we will take our variables of interest that will be used to train the AI model and organize them for ingestion into the machine learning model.