Overview¶
This notebooks shows how to query the Google Cloud CMIP6 catalog and load the data using Python.
Prerequisites¶
| Concepts | Importance | Notes |
|---|---|---|
| Intro to Xarray | Necessary | |
| Understanding of NetCDF | Helpful | Familiarity with metadata structure |
- Time to learn: 10 minutes
Imports¶
from matplotlib import pyplot as plt
import numpy as np
import pandas as pd
import xarray as xr
import zarr
import fsspec
import nc_time_axis
%matplotlib inline
plt.rcParams['figure.figsize'] = 12, 6Browse Catalog¶
The data catatalog is stored as a CSV file. Here we read it with Pandas.
df = pd.read_csv('https://storage.googleapis.com/cmip6/cmip6-zarr-consolidated-stores.csv')
df.head()The columns of the dataframe correspond to the CMI6 controlled vocabulary.
Here we filter the data to find monthly surface air temperature for historical experiments.
df_ta = df.query("activity_id=='CMIP' & table_id == 'Amon' & variable_id == 'tas' & experiment_id == 'historical'")
df_taNow we do further filtering to find just the models from NCAR.
df_ta_ncar = df_ta.query('institution_id == "NCAR"')
df_ta_ncarLoad Data¶
Now we will load a single store using fsspec, zarr, and xarray.
# get the path to a specific zarr store (the first one from the dataframe above)
zstore = df_ta_ncar.zstore.values[-1]
print(zstore)
# create a mutable-mapping-style interface to the store
mapper = fsspec.get_mapper(zstore)
# open it using xarray and zarr
ds = xr.open_zarr(mapper, consolidated=True)
dsPlot the Data¶
Plot a map from a specific date:
ds.tas.sel(time='1950-01').squeeze().plot()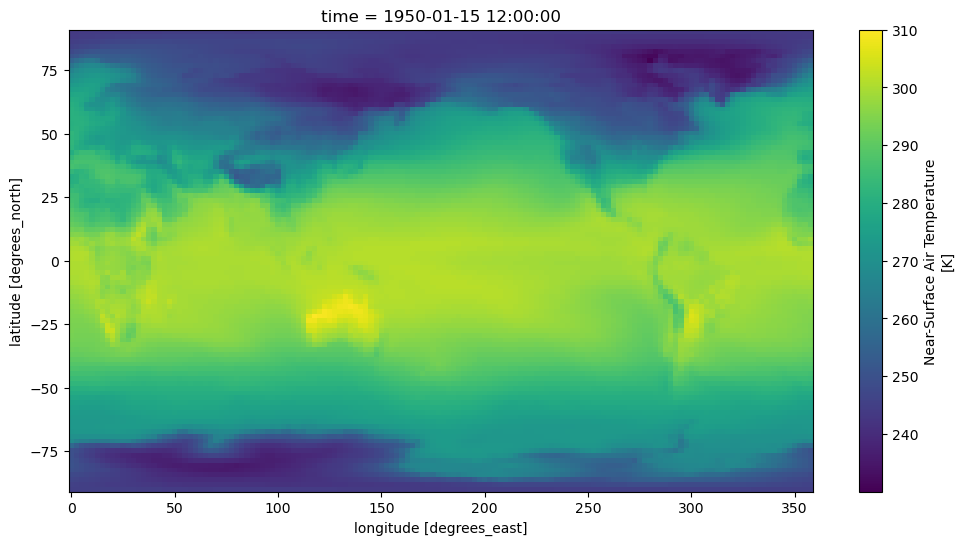
The global mean of a lat-lon field needs to be weighted by the area of each grid cell, which is proportional to the cosine of its latitude.
def global_mean(field):
weights = np.cos(np.deg2rad(field.lat))
return field.weighted(weights).mean(dim=['lat', 'lon'])We can pass all of the temperature data through this function:
ta_timeseries = global_mean(ds.tas)
ta_timeseriesBy default the data are loaded lazily, as Dask arrays. Here we trigger computation explicitly.
%time ta_timeseries.load()ta_timeseries.plot(label='monthly')
ta_timeseries.rolling(time=12).mean().plot(label='12 month rolling mean', color='k')
plt.legend()
plt.grid()
plt.title('Global Mean Surface Air Temperature')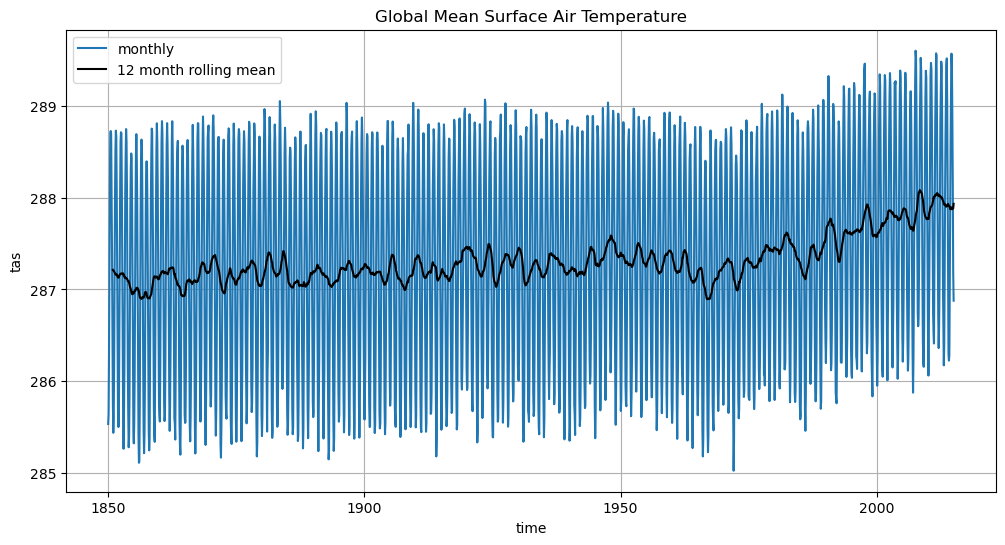
Summary¶
In this notebook, we opened a CESM2 dataset with fsspec and zarr. We calculated and plotted global average surface air temperature.
What’s next?¶
We will open a dataset with ESGF and OPenDAP.
Resources and references¶
- Original notebook in the Pangeo Gallery by Henri Drake and Ryan Abernathey