Exploratory Data Analysis
In this exercise, we are using machine learning as a tool for data exploration. We are interested in discovering the impact of the other attributes on PM10 dust concentration. One way to investigate this is to check the contribution of each attribute to the prediction accuracy of a learning algorithm. Our algorithm of chocie is random forests due to the interpretability of the resultant predictor.
# Importing relevant libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import sklearn
from sklearn.model_selection import cross_val_score
from sklearn.ensemble import RandomForestRegressor
Key packages
package |
Use |
|---|---|
Self-organizing maps (SOM) |
|
Machine learning |
|
for plotting |
Global figure settings
This piece of code sets the font size, line widths, figure title size, and resolution for all figures generated in this cookbook.
# customize figure
import matplotlib as mpl
mpl.rcParams['font.size'] = 28
mpl.rcParams['legend.fontsize'] = 'large'
mpl.rcParams['figure.titlesize'] = 'large'
mpl.rcParams['lines.linewidth'] = 2.5
mpl.rcParams['axes.linewidth'] = 2.5
mpl.rcParams["axes.unicode_minus"] = True
mpl.rcParams['figure.dpi'] = 150
mpl.rcParams['savefig.bbox']='tight'
mpl.rcParams['hatch.linewidth'] = 2.5
Import data
# Loading and visualizing the data
dust_df = pd.read_csv('../saharan_dust_met_vars.csv', index_col='time')
# print out shape of data
print('Shape of data:', np.shape(dust_df))
# print first 5 rows of data
print(dust_df.head())
Shape of data: (18466, 10)
PM10 T2 rh2 slp PBLH RAINC \
time
1960-01-01 2000.1490 288.24875 32.923786 1018.89420 484.91812 0.0
1960-01-02 4686.5370 288.88450 30.528862 1017.26575 601.58310 0.0
1960-01-03 5847.7515 290.97128 26.504536 1015.83514 582.38540 0.0
1960-01-04 5252.0586 292.20060 30.678936 1013.92230 555.11860 0.0
1960-01-05 3379.3190 293.06076 27.790462 1011.94934 394.95440 0.0
wind_speed_10m wind_speed_925hPa U10 V10
time
1960-01-01 6.801503 13.483623 -4.671345 -4.943579
1960-01-02 8.316340 18.027075 -6.334070 -5.388977
1960-01-03 9.148216 17.995173 -6.701636 -6.227193
1960-01-04 8.751743 15.806478 -6.387379 -5.982842
1960-01-05 6.393228 9.160809 -4.238991 -4.785845
What is the data distribution?
Understand your data first before usage.
plt.rcParams['figure.figsize'] = [22, 16]
dust_df.hist(bins=25);
plt.tight_layout()
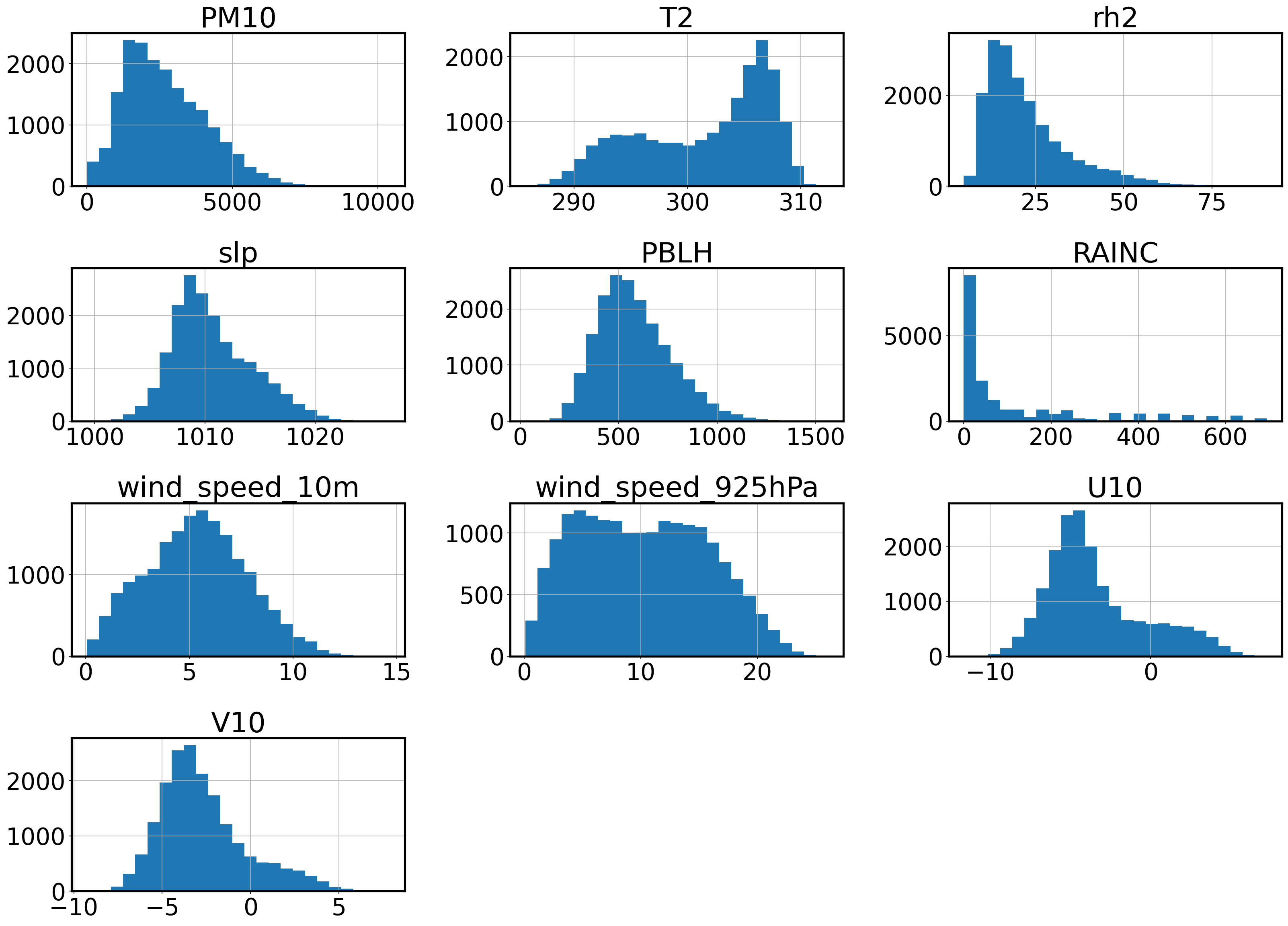
The histograms above are showing us the how the attributes vary across the samples in the dataset. The histogram that stands out the most is RAINC due to its strong concentration around a small section of its range. The long tail of the histogram extends to 700 to capture some rare occurences of heavy rain, but most days show little to no precipitation, falling under 100. In order to capture these rare events, the majority of the samples are compressed in a small range. To represent the typical days more accurately, we quantize RAINC to capture the rain events without the skewing effect of the concentrated distribution.
Using AMS definitions, and keeping in mind that our data captures a 24 hour range, we use the following conversion on the RAINC attribute:
0 –> No rain (value
0)(0, 24] –> Drizzle (value
1)(24,60] –> Light rain (value
2)(60,182] –> Moderate rain (value
3)182+ –> Heavy rain (value
4)
# Converting the RAINC attribute
def cat_precip(row):
if(row['RAINC'] == 0):
return 0 #'NR'
elif((row['RAINC'] > 0) and (row['RAINC'] <= 24)):
return 1 #'D'
elif((row['RAINC'] > 24) and (row['RAINC'] <= 60)):
return 2 #'LR'
elif((row['RAINC'] > 60) and (row['RAINC'] <= 182)):
return 3 #'MR'
elif(row['RAINC'] > 82):
return 4 #'HR'
dust_df['RAIN'] = dust_df.apply(cat_precip, axis=1)
dust_df = dust_df.drop(columns=["RAINC"])
Another factor in feature engineering is the over-representation of the wind speed. In adition to the wind speed attributes at two elevations (wind_speed_10m, wind_speed_925hPa), we have the U10 and V10 variables implicitly encoding the same information. We reduce this over-emphasis by converting the latter two variables into a categorical attribute that only encodes the wind direction in one of the 4 possible values: Southeast (SE), Southwest (SW), Northeast (NE) and Northwest (NW). In our dataset there is no instance where either of the directions are zero, so we don’t need to represent the directions East, West, South and North.
For further applications that qould require further detail in wind direction, one might consider increasing the granularity by including more categories like North-Northwest, West-Northwest, etc. that captures which of the two primary directions the wind is closer to.
# Converting U10 and V10 attributes
def cat_wind_dir(row):
if((row['U10']>=0) and (row['V10']>=0)):
return 'SW'
if((row['U10']>=0) and (row['V10']<0)):
return 'NW'
if((row['U10']<0) and (row['V10']>=0)):
return 'SE'
else:
return 'NE'
dust_df['WIND_DIR'] = dust_df.apply(cat_wind_dir, axis=1)
dust_df = dust_df.drop(columns=['U10','V10'])
dust_df.head()
| PM10 | T2 | rh2 | slp | PBLH | wind_speed_10m | wind_speed_925hPa | RAIN | WIND_DIR | |
|---|---|---|---|---|---|---|---|---|---|
| time | |||||||||
| 1960-01-01 | 2000.1490 | 288.24875 | 32.923786 | 1018.89420 | 484.91812 | 6.801503 | 13.483623 | 0 | NE |
| 1960-01-02 | 4686.5370 | 288.88450 | 30.528862 | 1017.26575 | 601.58310 | 8.316340 | 18.027075 | 0 | NE |
| 1960-01-03 | 5847.7515 | 290.97128 | 26.504536 | 1015.83514 | 582.38540 | 9.148216 | 17.995173 | 0 | NE |
| 1960-01-04 | 5252.0586 | 292.20060 | 30.678936 | 1013.92230 | 555.11860 | 8.751743 | 15.806478 | 0 | NE |
| 1960-01-05 | 3379.3190 | 293.06076 | 27.790462 | 1011.94934 | 394.95440 | 6.393228 | 9.160809 | 0 | NE |
NB: This dataframe of dust_df is saved on disk for future use.
Are there any outliers?
Some machine learning models do not perform well when there are outliers in the data. This section explores the data for any potential outliers. The boxplots show there are outliers in the data, hence we need to use a scaling method which is robust on outliers.
dust_df = pd.read_csv('../saharan_dust_met_vars.csv', index_col='time')
feature_names = dust_df.columns
fig, ax = plt.subplots(3,4, figsize=(26,12))
ax = ax.flatten()
for ind, col in enumerate(feature_names):
ax[ind].boxplot(dust_df[col].dropna(axis=0),
notch=True, whis=1.5,
showmeans=True)
ax[ind].grid(which='minor', axis='both')
ax[ind].set_xticklabels([''])
ax[ind].set_ylabel(col)
#ax[ind].set_title(col)
ax[ind].set_facecolor('white')
ax[ind].spines['bottom'].set_color('0.1')
ax[ind].spines['top'].set_color('0.1')
ax[ind].spines['right'].set_color('0.1')
ax[ind].spines['left'].set_color('0.1')
ax[ind].grid(True)
ax[10].set_axis_off()
ax[11].set_axis_off()
fig.tight_layout()
#plt.savefig('box_plots.png')
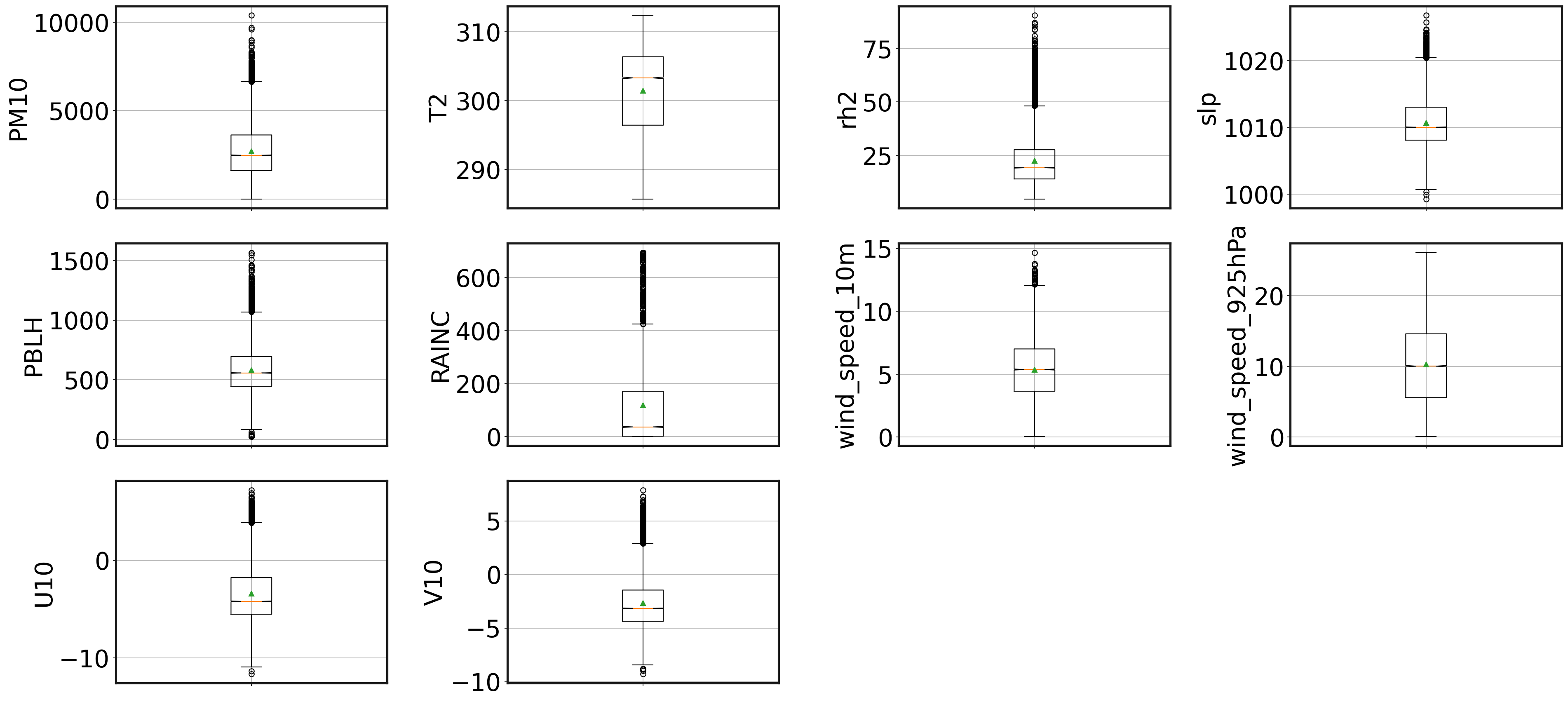
Scaling the variables
As you can see, there is a large range of values among variables. SOM and PCA are scale variant, so to not influence the results as it is the case in many unsupervised machine learning models, it is important to scale them. Many scaling methods exist, but we will use the robust scaling method since this takes care of outliers.
This concludes the primary exploration of our data. We now do a deeper dive into our exploration by using statistical analysis through Principal Component Analysis.
