# load python packages
import numpy as np
import pandas as pd
import xarray as xr
import math
import matplotlib as mpl
# load machine learning packages
import matplotlib.pyplot as plt
import sklearn
from sklearn import preprocessing
# minisom package for SOM
!pip install minisom
import minisom
from minisom import MiniSom
from matplotlib.patches import Patch
from matplotlib.patches import RegularPolygon, Ellipse
from mpl_toolkits.axes_grid1 import make_axes_locatable
from matplotlib import cm, colorbar
from matplotlib.lines import Line2D
import matplotlib.cbook as cbookRequirement already satisfied: minisom in /home/runner/micromamba/envs/dust-cookbook-dev/lib/python3.14/site-packages (2.3.6)
Global figure settings¶
# customize figure
mpl.rcParams['font.size'] = 18
mpl.rcParams['legend.fontsize'] = 'large'
mpl.rcParams['figure.titlesize'] = 'large'
mpl.rcParams['lines.linewidth'] = 2.5
mpl.rcParams['axes.linewidth'] = 2.5
mpl.rcParams["axes.unicode_minus"] = True
mpl.rcParams['figure.dpi'] = 150
mpl.rcParams['savefig.bbox']='tight'
mpl.rcParams['hatch.linewidth'] = 2.5# little function
def remove_time_mean(x):
return x - x.mean(dim='time')Importing data from disk¶
dust_df = pd.read_csv('../saharan_dust_met_vars.csv', index_col='time')
# print out shape of data
print('Shape of data:', np.shape(dust_df))
# print first 5 rows of data
print(dust_df.head())
feature_names = dust_df.columnsShape of data: (18466, 10)
PM10 T2 rh2 slp PBLH RAINC \
time
1960-01-01 2000.1490 288.24875 32.923786 1018.89420 484.91812 0.0
1960-01-02 4686.5370 288.88450 30.528862 1017.26575 601.58310 0.0
1960-01-03 5847.7515 290.97128 26.504536 1015.83514 582.38540 0.0
1960-01-04 5252.0586 292.20060 30.678936 1013.92230 555.11860 0.0
1960-01-05 3379.3190 293.06076 27.790462 1011.94934 394.95440 0.0
wind_speed_10m wind_speed_925hPa U10 V10
time
1960-01-01 6.801503 13.483623 -4.671345 -4.943579
1960-01-02 8.316340 18.027075 -6.334070 -5.388977
1960-01-03 9.148216 17.995173 -6.701636 -6.227193
1960-01-04 8.751743 15.806478 -6.387379 -5.982842
1960-01-05 6.393228 9.160809 -4.238991 -4.785845
Scaling the variables using the robust scaling method¶
# normalization by minmax scaling
minmax_sc = preprocessing.MinMaxScaler(feature_range = (0, 1))
minmax_sc.fit(dust_df)
minmax_scaled_df = minmax_sc.transform(dust_df)
# standardization
std_sc = preprocessing.StandardScaler().fit(dust_df)
std_scaled_df = std_sc.transform(dust_df)
# Robust scaling for outliers
rb_sc = preprocessing.RobustScaler().fit(dust_df)
rob_scaled_df = rb_sc.transform(dust_df)Define map and train the SOM map¶
# training data
train_data = rob_scaled_df
# Define minisom model
n_samples = train_data.shape[1] # retrieves the number of sampe (10 for this cookbook)
num = math.ceil(5*math.sqrt(n_samples))
som = MiniSom(x=num,
y=num, # map size, NxN
input_len=10, # number of features used for training the model (10 element input vectors)
sigma=3., # sigma: is the radius of the different neighbors in the SOM
learning_rate=0.5, # learning rate: determines how much the weights are adjusted during each iterations
neighborhood_function='gaussian', # a few options for this
topology='hexagonal',
activation_distance='euclidean',
random_seed=10)
# initilize weight
som.random_weights_init(train_data) # random weights initialization
#som.pca_weights_init(train_data) # initialize weights using PCA
## there are two type of training
# 1. train_random: trains model by pickinhg random data from the data
# 2. train_batch: trains model from samples in the order in which they are fed.
som.train(data = train_data, num_iteration = 25000,
verbose=True, random_order=True) # training the SOM model for 25000 iterations [ 18240 / 25000 ] 73% - 0:00:00 left [ 19777 / 25000 ] 79% - 0:00:00 left [ 21196 / 25000 ] 85% - 0:00:00 left [ 23196 / 25000 ] 93% - 0:00:00 left
quantization error: 0.7955823980825103
Visualizing the SOM results¶
Neighbour distance¶
The neighbor distance is also known as the “U-Matrix”. It is the distance between each node and its neighbours. Regions of low neighbourhood distance indicate groups of nodes that are similar, while regions of large distances indicate nodes are much more different. The U-Matrix can be used to identify clusters within the SOM map.
from pylab import bone, pcolor, colorbar, plot, show
bone()
pcolor(som.distance_map().T, cmap='bone')
colorbar()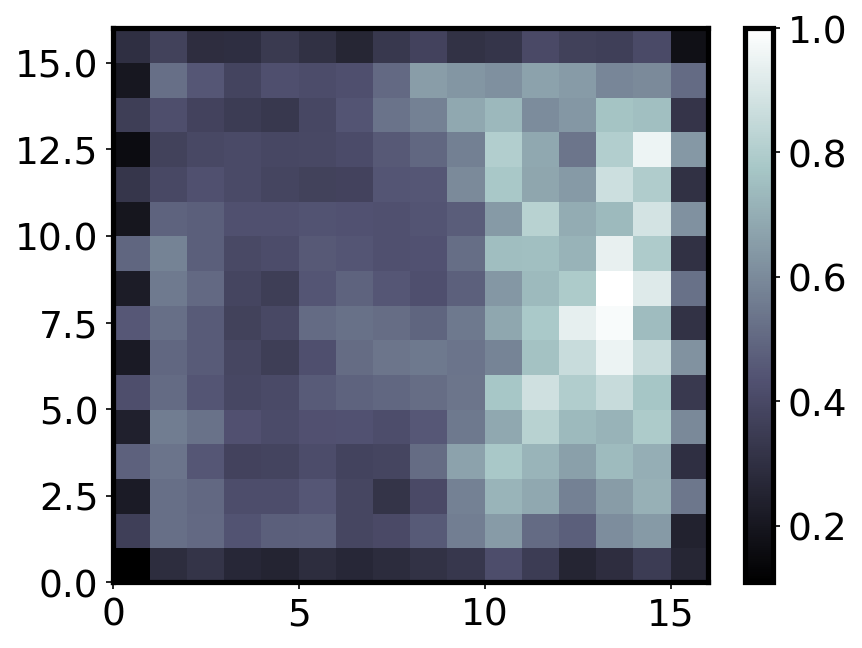
Feature plane¶
This is a map for each variable used, and it shows the magnitude of the weights associated to it for each neuron.
titles = ['PM10', '2m temperature', '2m relative humidity', 'Mean sea level pressure', 'Boundary layer depth',
'Convective rainfall', '10m wind speed', '925hPa wind speed', 'horizontal wind', 'meridional wind']
W = som.get_weights()
fig, ax = plt.subplots(3, 4, figsize=(16, 10))
ax = ax.flatten()
for i, f in enumerate(feature_names):
#plt.subplot(5, 2, i+1)
ax[i].set_title(titles[i])
ax[i].pcolor(W[:,:,i].T, cmap='coolwarm')
#ax[i].set_xticks(np.arange(num+1))
#ax[i].set_yticks(np.arange(num+1))
ax[10].set_axis_off()
ax[11].set_axis_off()
plt.tight_layout()
plt.savefig('feature_patterns.png')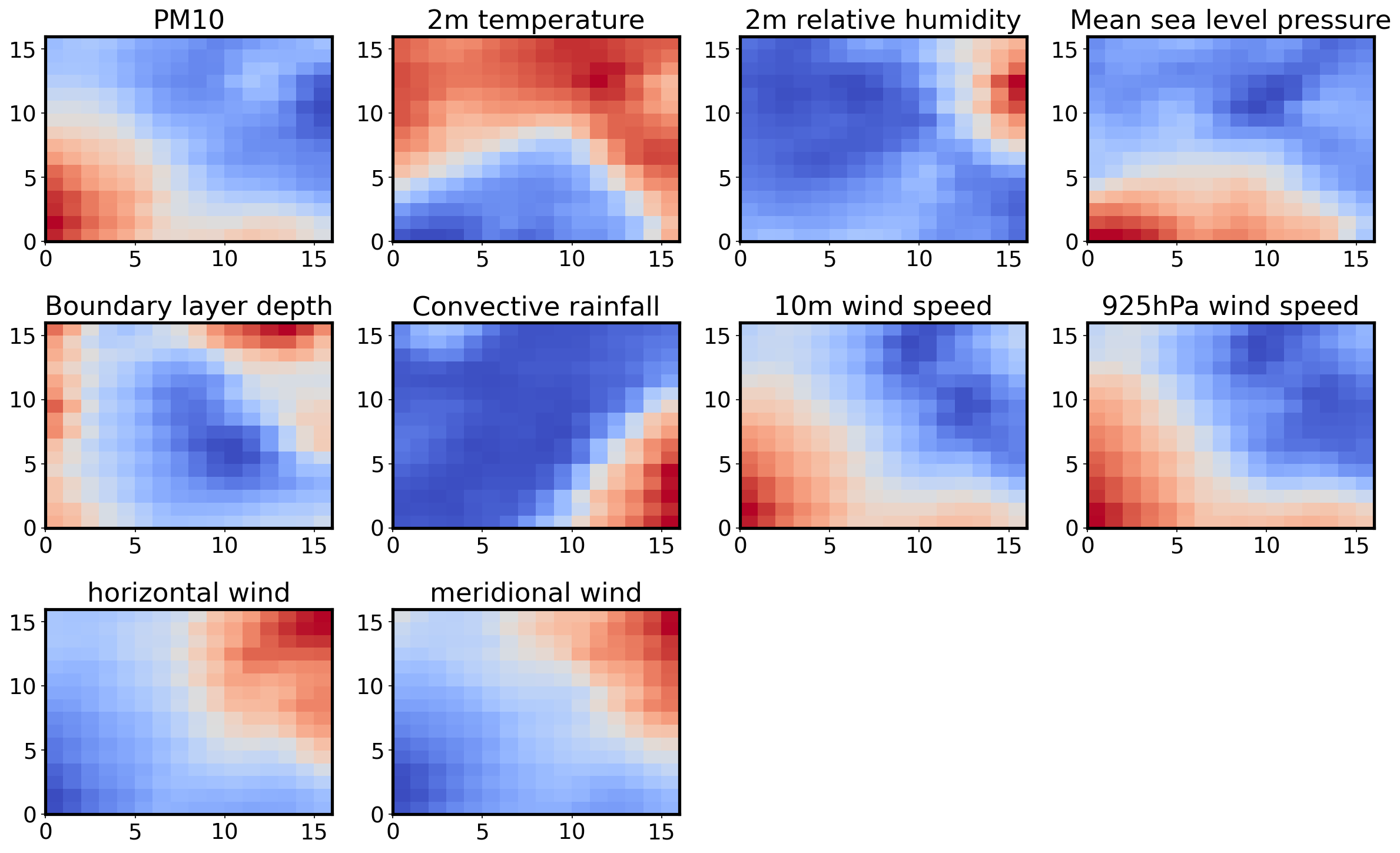
We see a pattern which establishes the relationship between each of the variables under consideration. For example, nodes of higher PM10 concentration are associated with nodes of higher 10m wind speed and 925hPa wind speed and vice versa. The horizontal and meridional wind patterns suggest a prevailing northeasterly trade winds, which are known to be associated with dust emissions and transport. Regions of high PM10 concentration correspond to somewhat lower temperatures, which may be expected, but more diurnal scale analysis is need to verify this statement. The analysis also reveals that increased dust concentration may lower relative humidity at the surface, and this could potentially affect tropical cyclone development over the North Atlantic as been speculated in many papers.
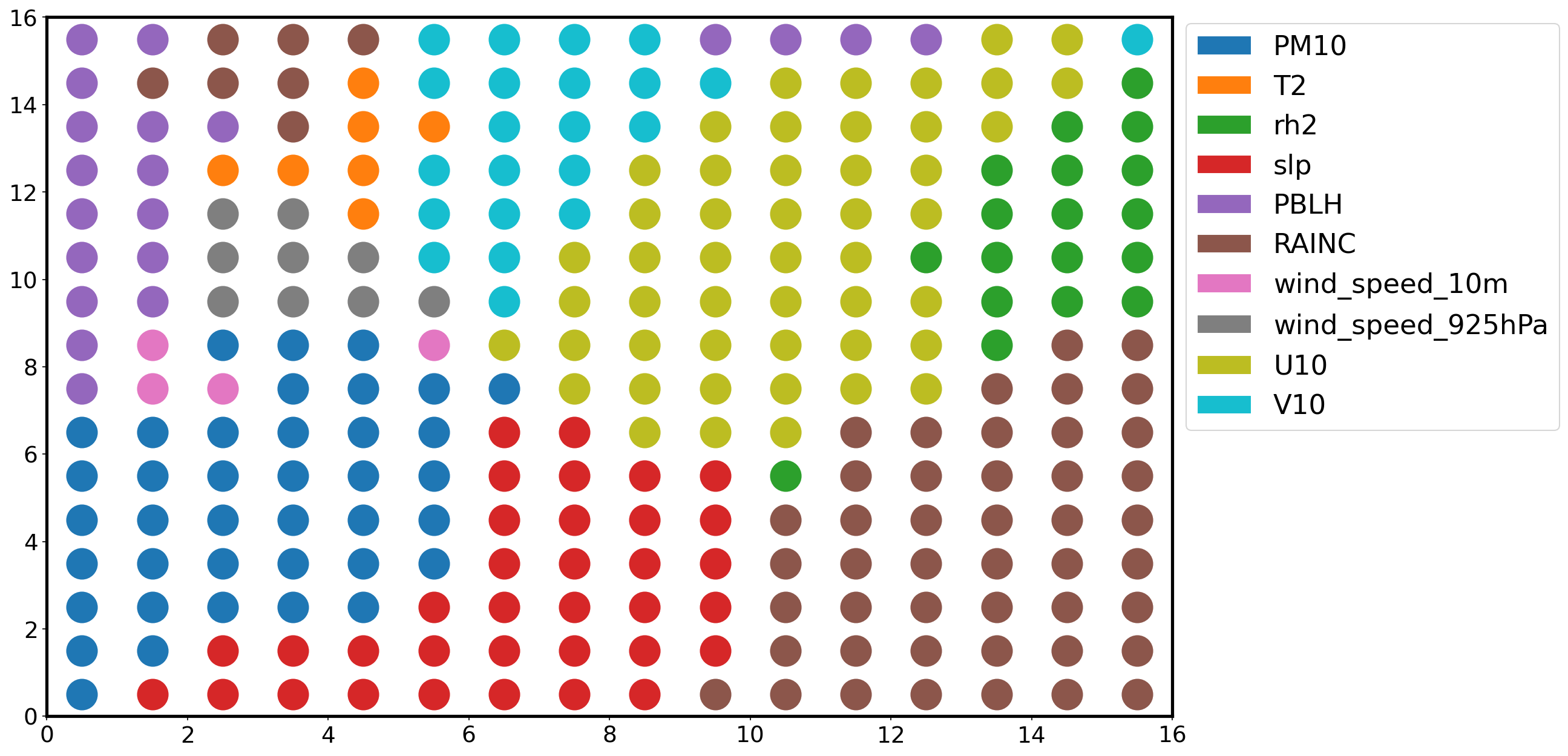
Most relevant feature plane¶
By associating each neuron to the feature with the maximum weight, we divide the map into distinct regions, where specific features are most important (high values).
Z = np.zeros((num, num))
W = som.get_weights()
plt.figure(figsize=(16, 10))
for i in np.arange(som._weights.shape[0]):
for j in np.arange(som._weights.shape[1]):
feature = np.argmax(W[i, j , :])
plt.plot([i+.5], [j+.5], color='C'+str(feature),
marker='o', markersize=24)
legend_elements = [Patch(facecolor='C'+str(i),
edgecolor='w',
label=f) for i, f in enumerate(feature_names)]
plt.legend(handles=legend_elements,
loc='center left',
bbox_to_anchor=(1, .7))
plt.xlim([0, num])
plt.ylim([0, num])
plt.show()