Introduction to Verde
Unlike xESMF, verde is mainly designed around unstructured grids. Verde has a lot of great examples, we will use an atmospheric one below, but highly reccomend reading though the documents to get a better handle on the API.
https://www.fatiando.org/verde/latest/
Prerequisites
Knowing your way around pandas, xarray, numpy and matplotlib is beneficial. This is not deisgned to be an introduction to any of those packages. We will be using Cartopy for some of the plots as well.
Imports
import pandas as pd
import numpy as np
import cartopy.crs as ccrs
import cartopy.feature as cf
from appdirs import *
import verde as vd
import matplotlib.pyplot as plt
import matplotlib.ticker as mticker
%load_ext watermark
%watermark --iversions
cartopy : 0.23.0
verde : 1.8.0
sys : 3.10.14 | packaged by conda-forge | (main, Mar 20 2024, 12:45:18) [GCC 12.3.0]
matplotlib: 3.8.4
numpy : 1.26.4
pandas : 2.2.2
colnames=['remove', 'lon', 'lat', 'date', 'sensor', 'PM25value', 'sensor_type']
df = pd.read_csv('../data/airnow_data.csv', names=colnames, header=None)
df = df.drop(columns='remove')
df.head(3)
| lon | lat | date | sensor | PM25value | sensor_type | |
|---|---|---|---|---|---|---|
| 0 | 41.783083 | -110.537888 | 2022-09-05T16:00 | PM2.5 | 45 | 1 |
| 1 | 40.294300 | -110.009000 | 2022-09-05T16:00 | PM2.5 | 40 | 1 |
| 2 | 40.464722 | -109.560830 | 2022-09-05T16:00 | PM2.5 | 30 | 1 |
df.date = pd.to_datetime(df.date)
df.dtypes
lon float64
lat float64
date datetime64[ns]
sensor object
PM25value int64
sensor_type int64
dtype: object
Let’s pick a time that has decent coverage:
df.date.value_counts().head()
date
2022-09-05 16:00:00 36
2022-09-06 07:00:00 36
2022-09-06 20:00:00 36
2022-09-06 19:00:00 36
2022-09-06 18:00:00 36
Name: count, dtype: int64
df2 = df[df.date == '2022-09-06T07:00']
df2.head(3)
| lon | lat | date | sensor | PM25value | sensor_type | |
|---|---|---|---|---|---|---|
| 540 | 41.783083 | -110.537888 | 2022-09-06 07:00:00 | PM2.5 | 16 | 1 |
| 541 | 40.294300 | -110.009000 | 2022-09-06 07:00:00 | PM2.5 | 29 | 1 |
| 542 | 40.464722 | -109.560830 | 2022-09-06 07:00:00 | PM2.5 | 23 | 1 |
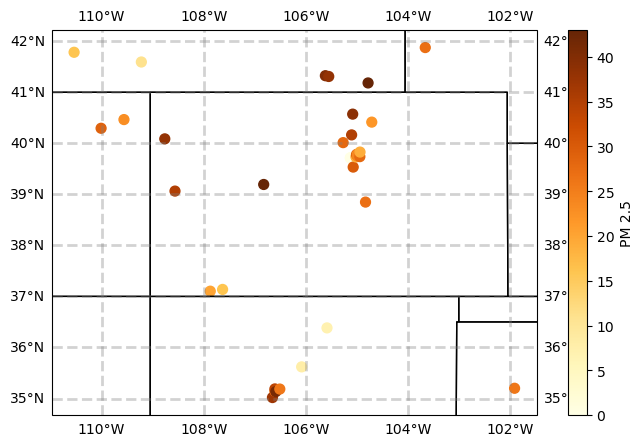
Plotting the data
plt.figure(figsize=(8, 5))
ax = plt.axes(projection=ccrs.PlateCarree())
ax.add_feature(cf.STATES)
gl = ax.gridlines(crs=ccrs.PlateCarree(), draw_labels=True,
linewidth=2, color='gray', alpha=0.35, linestyle='--')
gl.xlabels_top = False
gl.ylabels_right = False
gl.xlocator = mticker.FixedLocator([-110, -108, -106, -104, -102])
plt.scatter(df2.lat,
df2.lon,
c = df2.PM25value,
s=50,
cmap="YlOrBr")
plt.colorbar().set_label("PM 2.5")
plt.show()
/home/runner/miniconda3/envs/gridding-cookbook-dev/lib/python3.10/site-packages/cartopy/io/__init__.py:241: DownloadWarning: Downloading: https://naturalearth.s3.amazonaws.com/10m_cultural/ne_10m_admin_1_states_provinces_lakes.zip
warnings.warn(f'Downloading: {url}', DownloadWarning)
Verde Workflows
Some of these workflows might not work super well from a scientific standpoint, this is just to show the mechanics of the verde package.
coordinates = (df2.lon, df2.lat)
Trend Estimation
Same code, different example can be found here: https://www.fatiando.org/verde/latest/tutorials/trends.html
trend = vd.Trend(degree=1).fit(coordinates, df2.PM25value)
print(trend.coef_)
[38.39163129 0.54379964 0.32281663]
/home/runner/miniconda3/envs/gridding-cookbook-dev/lib/python3.10/site-packages/verde/base/least_squares.py:68: FutureWarning: Series.ravel is deprecated. The underlying array is already 1D, so ravel is not necessary. Use `to_numpy()` for conversion to a numpy array instead.
regr.fit(jacobian, data.ravel(), sample_weight=weights)
trend_values = trend.predict(coordinates)
residuals = df2.PM25value - trend_values
plt.figure(figsize=(8, 5))
ax = plt.axes(projection=ccrs.PlateCarree())
ax.add_feature(cf.STATES)
gl = ax.gridlines(crs=ccrs.PlateCarree(), draw_labels=True,
linewidth=2, color='gray', alpha=0.35, linestyle='--')
gl.top_labels = False
gl.ylabels_right = False
gl.xlocator = mticker.FixedLocator([-110, -108, -106, -104, -102])
maxabs = vd.maxabs(residuals)
plt.scatter(df2.lat,
df2.lon,
c = residuals,
s=50,
vmin=-maxabs,
vmax=maxabs,
cmap="bwr")
plt.colorbar().set_label("Residuals")
plt.show()
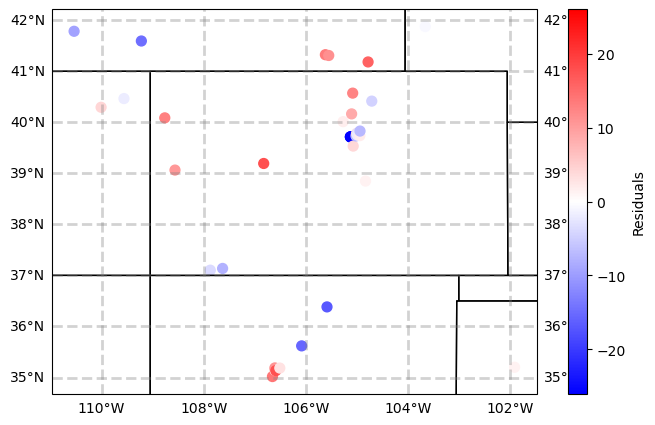
Unlike the Texas example in Verde, hard to see a distinct regional trend.
Data Decimation
spacing = 30 / 60 # Play around with this!
reducer = vd.BlockReduce(reduction=np.median, spacing= spacing)
print(reducer)
BlockReduce(reduction=<function median at 0x7fb078359770>, spacing=0.5)
filter_coords, filter_data = reducer.filter(
coordinates=(df2.lat, df2.lon), data=df2.PM25value)
/home/runner/miniconda3/envs/gridding-cookbook-dev/lib/python3.10/site-packages/verde/blockreduce.py:177: FutureWarning: Series.ravel is deprecated. The underlying array is already 1D, so ravel is not necessary. Use `to_numpy()` for conversion to a numpy array instead.
columns = {"data{}".format(i): comp.ravel() for i, comp in enumerate(data)}
/home/runner/miniconda3/envs/gridding-cookbook-dev/lib/python3.10/site-packages/verde/blockreduce.py:179: FutureWarning: The provided callable <function median at 0x7fb078552ef0> is currently using DataFrameGroupBy.median. In a future version of pandas, the provided callable will be used directly. To keep current behavior pass the string "median" instead.
blocked = pd.DataFrame(columns).groupby("block").aggregate(reduction)
/home/runner/miniconda3/envs/gridding-cookbook-dev/lib/python3.10/site-packages/verde/blockreduce.py:231: FutureWarning: Series.ravel is deprecated. The underlying array is already 1D, so ravel is not necessary. Use `to_numpy()` for conversion to a numpy array instead.
"coordinate{}".format(i): coord.ravel()
/home/runner/miniconda3/envs/gridding-cookbook-dev/lib/python3.10/site-packages/verde/blockreduce.py:236: FutureWarning: The provided callable <function median at 0x7fb078552ef0> is currently using DataFrameGroupBy.median. In a future version of pandas, the provided callable will be used directly. To keep current behavior pass the string "median" instead.
grouped = table.groupby("block").aggregate(self.reduction)
Sanity check
np.shape(filter_coords)[1] == np.size(filter_data)
True
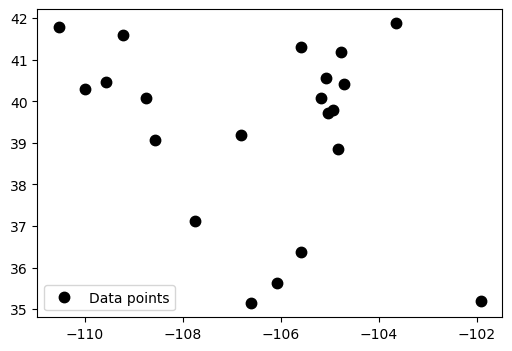
Plotting the decimated datapoints
plt.figure(figsize=(6, 4))
plt.plot(*filter_coords, ".k", markersize=15, label='Data points')
plt.legend()
plt.show()
Verde Spline
This turtorial runs though this entire workflow, but with a bathymetery dataset: https://www.fatiando.org/verde/latest/tutorials/projections.html
spline = vd.Spline().fit(filter_coords, filter_data)
region = vd.get_region(filter_coords)
grid_coords = vd.grid_coordinates(region, spacing=spacing)
grid = spline.grid(coordinates=grid_coords, data_names="PM25")
print(grid)
<xarray.Dataset> Size: 2kB
Dimensions: (northing: 14, easting: 18)
Coordinates:
* easting (easting) float64 144B -110.5 -110.0 -109.5 ... -102.4 -101.9
* northing (northing) float64 112B 35.14 35.66 36.18 ... 40.84 41.36 41.88
Data variables:
PM25 (northing, easting) float64 2kB 74.69 69.09 63.62 ... 11.73 6.292
Attributes:
metadata: Generated by Spline(mindist=0)
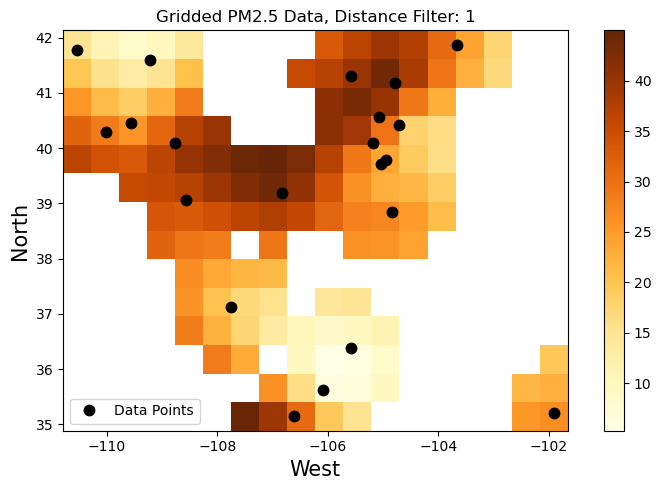
distance_mask = 1 #this might not make physical sense
grid = vd.distance_mask(filter_coords, maxdist=distance_mask, grid=grid)
Let’s make a plot with the gridded and masked output, also shows where the datapoints are:
plt.figure(figsize=(8, 5))
pc = grid.PM25.plot.pcolormesh(cmap="YlOrBr", add_colorbar=False)
plt.colorbar(pc)
plt.plot(filter_coords[0], filter_coords[1], ".k", markersize=15, label='Data Points')
plt.title('Gridded PM2.5 Data, Distance Filter: '+str(distance_mask))
plt.xlabel("West", size=15)
plt.ylabel("North", size=15)
plt.legend()
plt.gca().set_aspect("equal")
plt.tight_layout()
plt.show()
Xarray
Making the grid into an xarray dataset is 1 line of code!
ds = grid.PM25.to_dataset()
ds
<xarray.Dataset> Size: 2kB
Dimensions: (easting: 18, northing: 14)
Coordinates:
* easting (easting) float64 144B -110.5 -110.0 -109.5 ... -102.4 -101.9
* northing (northing) float64 112B 35.14 35.66 36.18 ... 40.84 41.36 41.88
Data variables:
PM25 (northing, easting) float64 2kB nan nan nan nan ... 17.54 nan nanExporting this as a netCDF:
ds.to_netcdf('test.nc')
Summary
Verde and xESMF are two different gridding packages, with two different aims. They both work well with xarray!
