xESMF Demo using SEVIER dataset for Machine Learning Preperation
Thomas Martin @ThomasMGeo
Sept 2 - 2022
Overview
This is a quick (~20 minute) notebook that will cover how to use xESMF to regrid with xarray on a dataset. This notebooks will heavily borrow from this repo: https://github.com/ai2es/WAF_ML_Tutorial_Part1 & this paper https://proceedings.neurips.cc/paper/2020/file/fa78a16157fed00d7a80515818432169-Paper.pdf . Randy Chase (@dopplershift on github) is thanked for dataset prep and previous work!
Prerequisits
Working knowldge of xarray, matplotlib, and numpy is beneficial. This is not designed to be an introduction to any of those packages.
Imports
import numpy as np
import xarray as xr
import xesmf as xe
# Plotting utilities
import seaborn as sns
import matplotlib.pyplot as plt
Watermark is great repo to track versions when sharing work with notebooks. xESMF can be a little tricky to install, highly reccomend to install via conda instread of pip.
%load_ext watermark
%watermark --iversions
numpy : 1.26.4
matplotlib: 3.8.4
xesmf : 0.8.5
seaborn : 0.13.2
xarray : 2024.5.0
Dataset Load
file = '../data/onestorm.nc' # netcdf file
#open an example storm
ds = xr.open_dataset(file)
#see the data by printing ds. By putting at the bottom of the cell, it is automatically printed
ds
<xarray.Dataset> Size: 39MB
Dimensions: (x: 768, y: 768, t: 12, x2: 192, y2: 192, x3: 384,
y3: 384, x4: 48, y4: 48)
Dimensions without coordinates: x, y, t, x2, y2, x3, y3, x4, y4
Data variables:
visible (x, y, t) float32 28MB ...
water_vapor (x2, y2, t) float32 2MB ...
clean_infrared (x2, y2, t) float32 2MB ...
vil (x3, y3, t) float32 7MB ...
lightning_flashes (x4, y4, t) float32 111kB ...
Attributes:
t: time dimension of all images. These are 5-min time steps
x: x pixel dimension of the visible imagery
y: y pixel dimension of the visible imagery
x2: x pixel dimension of the water vapor and infrared imagery
y2: y pixel dimension the water vapor and infrared imagery
x3: x pixel dimension of the vertically integrated liquid imagery
y3: y pixel dimension the vertically integrated liquid imagery
x4: x pixel dimension of the lightning flashes
y4: y pixel dimension of the lightning flashesWe will be using this single dataset for the entirety of the notebook
plt.figure(figsize=(9,6))
#show all x pixels (:) and all y pixels (:) and the first time step, with a Grey colorscale, and the color min 0 and color max 1.
plt.imshow(ds.visible.isel(t=0)[:,:]*1e-4,cmap='Greys_r',vmin=0,vmax=1) # At timestep 0
#show us the colorbar
plt.colorbar(label='Visible Reflectance Factor')
#a function that cleans some of the figure up.
plt.tight_layout()
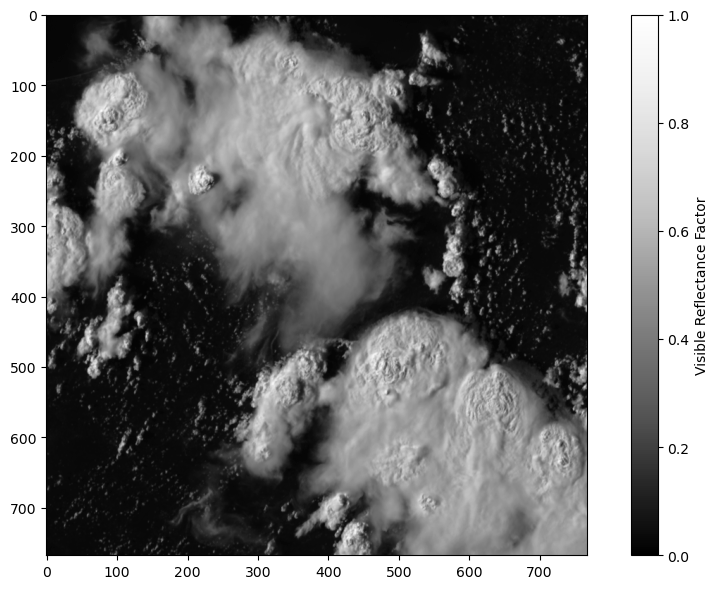
Let’s Zoom in one one patch, to gain some intuition on how satellite data is used in Machine Learning (ML)
# Bounds on Zoom box
xmin = 278
xmax = 288
ymin= 278
ymax= 288
plt.figure(figsize=(10,9))
#show all x pixels (:) and all y pixels (:) and the first time step, with a blue colorscale, and the color min 0 and color max 1.
sns.heatmap(ds.visible[xmin:xmax,ymin:ymax,0]*1e-4,
cmap="Blues",
annot=True,
annot_kws={"size": 11}, # font size
vmin=0,
vmax=1)
plt.show()
plt.tight_layout()
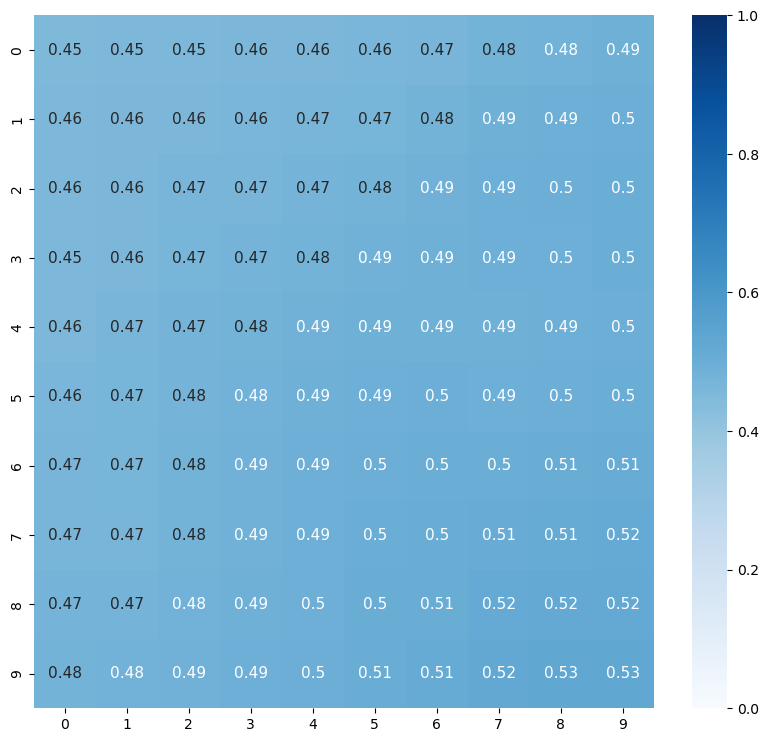
<Figure size 640x480 with 0 Axes>
Let’s check out a small section of the lightning strike data:
plt.figure(figsize=(6,5))
sns.heatmap(ds.lightning_flashes.isel(t=0)[3:9,3:9],
cmap="Blues",
annot=True,
annot_kws={"size": 11}) # font size
plt.show()
plt.tight_layout()
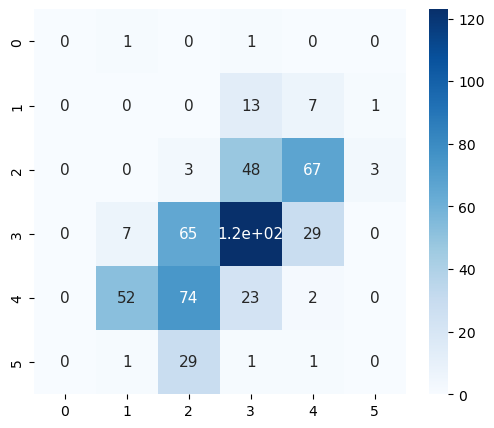
<Figure size 640x480 with 0 Axes>
After reviewing the dataset, some of the variables have different shapes:
print(ds.visible.shape) # x, y, time
print(ds.lightning_flashes.shape)
(768, 768, 12)
(48, 48, 12)
Using xESMF to regrid an xarray dataset:
Information available here: https://xesmf.readthedocs.io/en/latest/notebooks/Rectilinear_grid.html
In order to re-grid, let’s set up some coordinates. Lets do some sanity checking to check what the axises needs to be divided by:
# Why 47 and not 48? The range for x4 is 0 to 47 (48 steps). If you divide by 48, it will be longer by ~1 in each dimmension
np.shape(ds.visible.values)[0]/47 # for both x & y, this number does not need to be an integer
16.340425531914892
scaling_factor = np.shape(ds.lightning_flashes.values)
ds2 = ds.assign_coords(x4=ds.x4, y4=ds.y4,
x=ds.x/16.3404, y=ds.y/16.340425, # make the coordinates match
time=ds.t) # making it a new dataset
ds2
<xarray.Dataset> Size: 39MB
Dimensions: (x: 768, y: 768, t: 12, x2: 192, y2: 192, x3: 384,
y3: 384, x4: 48, y4: 48)
Coordinates:
* x4 (x4) int64 384B 0 1 2 3 4 5 6 7 ... 41 42 43 44 45 46 47
* y4 (y4) int64 384B 0 1 2 3 4 5 6 7 ... 41 42 43 44 45 46 47
* x (x) float64 6kB 0.0 0.0612 0.1224 ... 46.82 46.88 46.94
* y (y) float64 6kB 0.0 0.0612 0.1224 ... 46.82 46.88 46.94
time (t) int64 96B 0 1 2 3 4 5 6 7 8 9 10 11
Dimensions without coordinates: t, x2, y2, x3, y3
Data variables:
visible (x, y, t) float32 28MB 242.0 234.0 ... 3.238e+03
water_vapor (x2, y2, t) float32 2MB ...
clean_infrared (x2, y2, t) float32 2MB ...
vil (x3, y3, t) float32 7MB ...
lightning_flashes (x4, y4, t) float32 111kB 0.0 0.0 0.0 0.0 ... 0.0 0.0 0.0
Attributes:
t: time dimension of all images. These are 5-min time steps
x: x pixel dimension of the visible imagery
y: y pixel dimension of the visible imagery
x2: x pixel dimension of the water vapor and infrared imagery
y2: y pixel dimension the water vapor and infrared imagery
x3: x pixel dimension of the vertically integrated liquid imagery
y3: y pixel dimension the vertically integrated liquid imagery
x4: x pixel dimension of the lightning flashes
y4: y pixel dimension of the lightning flashesDecreasing visible resoultion to lighting resoultion
Making new datasets, this might not be required.
ds_visible = ds2["visible"].to_dataset()
ds_visible = ds_visible.rename({'x': 'lon','y': 'lat'})
ds_visible
<xarray.Dataset> Size: 28MB
Dimensions: (lon: 768, lat: 768, t: 12)
Coordinates:
* lon (lon) float64 6kB 0.0 0.0612 0.1224 0.1836 ... 46.82 46.88 46.94
* lat (lat) float64 6kB 0.0 0.0612 0.1224 0.1836 ... 46.82 46.88 46.94
time (t) int64 96B 0 1 2 3 4 5 6 7 8 9 10 11
Dimensions without coordinates: t
Data variables:
visible (lon, lat, t) float32 28MB 242.0 234.0 ... 3.517e+03 3.238e+03ds_lf = ds2["lightning_flashes"].to_dataset()
ds_lf = ds_lf.rename({'x4': 'lon',
'y4': 'lat'})
ds_lf
<xarray.Dataset> Size: 111kB
Dimensions: (lon: 48, lat: 48, t: 12)
Coordinates:
* lon (lon) int64 384B 0 1 2 3 4 5 6 7 ... 41 42 43 44 45 46 47
* lat (lat) int64 384B 0 1 2 3 4 5 6 7 ... 41 42 43 44 45 46 47
time (t) int64 96B 0 1 2 3 4 5 6 7 8 9 10 11
Dimensions without coordinates: t
Data variables:
lightning_flashes (lon, lat, t) float32 111kB 0.0 0.0 0.0 ... 0.0 0.0 0.0regridder = xe.Regridder(ds_visible, ds_lf, "bilinear")
regridder # print basic regridder information.
xESMF Regridder
Regridding algorithm: bilinear
Weight filename: bilinear_768x768_48x48.nc
Reuse pre-computed weights? False
Input grid shape: (768, 768)
Output grid shape: (48, 48)
Periodic in longitude? False
dr_out = regridder(ds_visible)
dr_out
/home/runner/miniconda3/envs/gridding-cookbook-dev/lib/python3.10/site-packages/xesmf/smm.py:131: UserWarning: Input array is not C_CONTIGUOUS. Will affect performance.
warnings.warn('Input array is not C_CONTIGUOUS. ' 'Will affect performance.')
<xarray.Dataset> Size: 111kB
Dimensions: (t: 12, lat: 48, lon: 48)
Coordinates:
time (t) int64 96B 0 1 2 3 4 5 6 7 8 9 10 11
* lon (lon) int64 384B 0 1 2 3 4 5 6 7 8 9 ... 39 40 41 42 43 44 45 46 47
* lat (lat) int64 384B 0 1 2 3 4 5 6 7 8 9 ... 39 40 41 42 43 44 45 46 47
Dimensions without coordinates: t
Data variables:
visible (t, lat, lon) float32 111kB 242.0 241.0 241.0 235.1 ... 0.0 0.0 0.0
Attributes:
regrid_method: bilinearDirect Comparison
Note the difference in x & y axis
time_step = 0
f, (ax1, ax2) = plt.subplots(1, 2, sharey=False, figsize=(8,8))
# Original figure
ax1.imshow(ds.visible.isel(t=time_step)[:,:]*1e-4,cmap='Greys_r',vmin=0,vmax=1) # the x and y axis were flipped, added the .T to fix the plot
ax1.title.set_text('Original')
ax2.imshow(dr_out.visible.isel(t=time_step)[:,:].T*1e-4,cmap='Greys_r',vmin=0,vmax=1)
ax2.title.set_text('Downscaled - Bilinear')
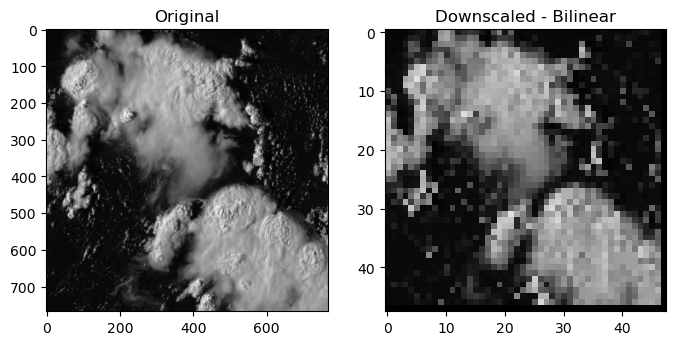
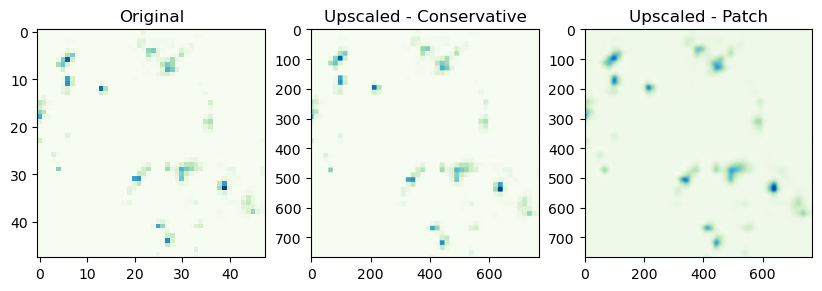
We can also upscale low-resoultion data
Note: This is not a reccomendation to do this for every workflow/dataset! There are five different algorithms that you can use, here is a nice comparison: https://xesmf.readthedocs.io/en/latest/notebooks/Compare_algorithms.html
regridder_up_con = xe.Regridder(ds_lf, ds_visible, "conservative") #note different method from before
regridder_up_patch = xe.Regridder(ds_lf, ds_visible, "patch")
regridder_up_patch # print basic regridder information.
xESMF Regridder
Regridding algorithm: patch
Weight filename: patch_48x48_768x768.nc
Reuse pre-computed weights? False
Input grid shape: (48, 48)
Output grid shape: (768, 768)
Periodic in longitude? False
upscaled_lf_con = regridder_up_con(ds_lf)
upscaled_lf_patch = regridder_up_patch(ds_lf)
/home/runner/miniconda3/envs/gridding-cookbook-dev/lib/python3.10/site-packages/xesmf/smm.py:131: UserWarning: Input array is not C_CONTIGUOUS. Will affect performance.
warnings.warn('Input array is not C_CONTIGUOUS. ' 'Will affect performance.')
/home/runner/miniconda3/envs/gridding-cookbook-dev/lib/python3.10/site-packages/xesmf/smm.py:131: UserWarning: Input array is not C_CONTIGUOUS. Will affect performance.
warnings.warn('Input array is not C_CONTIGUOUS. ' 'Will affect performance.')
f, (ax1, ax2, ax3) = plt.subplots(1, 3, sharey=False, figsize=(10,4))
# Original Dataset
ax1.imshow(ds.lightning_flashes.isel(t=0)[:,:],cmap='GnBu')
ax1.title.set_text('Original')
ax2.imshow(upscaled_lf_con.lightning_flashes.isel(t=0)[:,:].T, cmap='GnBu')
ax2.title.set_text('Upscaled - Conservative')
ax3.imshow(upscaled_lf_patch.lightning_flashes.isel(t=0)[:,:].T, cmap='GnBu')
ax3.title.set_text('Upscaled - Patch')
Merging Xarray Datasets
After you have made a new grid, you will want to combine them into a new xarray dataset for future analysis. Here is an example using combine by coords:
upscaled_lf_patch
<xarray.Dataset> Size: 28MB
Dimensions: (t: 12, lat: 768, lon: 768)
Coordinates:
time (t) int64 96B 0 1 2 3 4 5 6 7 8 9 10 11
* lon (lon) float64 6kB 0.0 0.0612 0.1224 ... 46.82 46.88 46.94
* lat (lat) float64 6kB 0.0 0.0612 0.1224 ... 46.82 46.88 46.94
Dimensions without coordinates: t
Data variables:
lightning_flashes (t, lat, lon) float32 28MB 0.0 0.0 0.0 ... 0.0 0.0 0.0
Attributes:
regrid_method: patchds_visible
<xarray.Dataset> Size: 28MB
Dimensions: (lon: 768, lat: 768, t: 12)
Coordinates:
* lon (lon) float64 6kB 0.0 0.0612 0.1224 0.1836 ... 46.82 46.88 46.94
* lat (lat) float64 6kB 0.0 0.0612 0.1224 0.1836 ... 46.82 46.88 46.94
time (t) int64 96B 0 1 2 3 4 5 6 7 8 9 10 11
Dimensions without coordinates: t
Data variables:
visible (lon, lat, t) float32 28MB 242.0 234.0 ... 3.517e+03 3.238e+03new_dataset = xr.combine_by_coords([upscaled_lf_patch, ds_visible])
new_dataset
<xarray.Dataset> Size: 57MB
Dimensions: (t: 12, lat: 768, lon: 768)
Coordinates:
time (t) int64 96B 0 1 2 3 4 5 6 7 8 9 10 11
* lon (lon) float64 6kB 0.0 0.0612 0.1224 ... 46.82 46.88 46.94
* lat (lat) float64 6kB 0.0 0.0612 0.1224 ... 46.82 46.88 46.94
Dimensions without coordinates: t
Data variables:
lightning_flashes (t, lat, lon) float32 28MB 0.0 0.0 0.0 ... 0.0 0.0 0.0
visible (lon, lat, t) float32 28MB 242.0 234.0 ... 3.238e+03
Attributes:
regrid_method: patchSummary
This has been a quick introduction to useing the xESMF regridding tools! The documentation for xESMF & xarray are very helpful for future learning.
