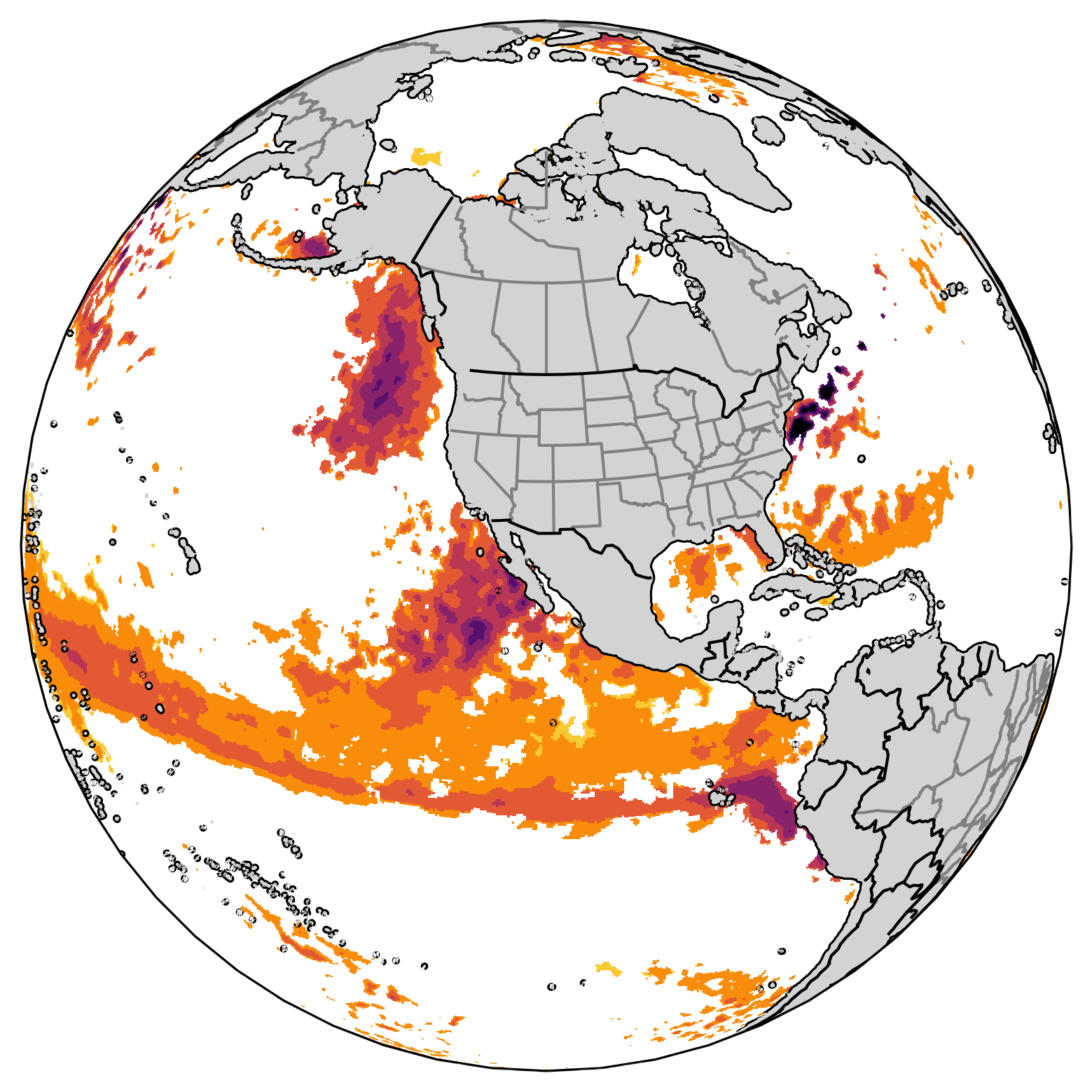
Marine Heatwave Detection¶
Marine heatwaves are periods of persistent anomalously warm ocean temperatures, which can have significant impacts on marine life as well as coastal communities and economies. To detect the warm ocean water, sea surface temperature (SST) is usually used to define if there is any marine heatwave event. The following example is following the paper Jacox et al., 2022
Overview¶
In this page/notebook, we will be go throught the following steps
Extract the data from the PSL OPeNDAP server
Calculate the SST climatology
Calculate the SST anomaly
Determine the SST threshold based on the anomaly
Identify the marine heatwaves based on threshold
Prerequisites¶
To better understand and follow the steps in the notebook, it will be helpful for user to go through
| Concepts | Importance | Notes |
|---|---|---|
| Xarray | Helpful | Chunking and OPeNDAP access |
Time to learn: 15 minutes.
System requirements:
python
Xarray
pydap (not imported but will be used in the Xarray backend)
matplotlib (not imported but will be used in the Xarray plotting)
Numpy
plotly (only for the final interactive plot)
Imports¶
# import the needed packages
import warnings
import xarray as xr
import numpy as np
import plotly.express as pxwarnings.simplefilter("ignore")warnings.simplefilter
This line of code is not affecting the execution but just removing some of the warning output that might clutter your notebook. However, do pay attention to some of the warnings since they will indicate some deprecation of function or arg/kwarg in future update.Extract the data from an OPeNDAP server¶
In this page/notebook, we demonstrate how to use the NOAA OISST v2 High-resolution dataset to detect marine heatwaves. The dataset is currently hosted by NOAA Physical Sciences Laboratory.
Info
To explore more gridded datasets that are hosted at NOAA PSL, here is a useful search toolopendap_mon_url = "https://psl.noaa.gov/thredds/dodsC/Datasets/noaa.oisst.v2.highres/sst.mon.mean.nc"Xarray getting remote data¶
Xarray has a great support on accessing data in the cloud.
It has been continue to expend its capability and functionality with the community discussion like this.
Here we use the xr.open_dataset method with the keyword argument (engine='pydap') to use the pydap package in the backend to access the OPeNDAP server.
ds_mon = xr.open_dataset(opendap_mon_url, engine='pydap', chunks={'time':12,'lon':-1,'lat':-1})Lazy Loading
We can load the data lazily (only loading the metadata and coordinates information) and peek at the data's dimension and availability on our local machine. The actual data (SST values at each grid point in this case) will only be downloaded from the PSL server when further data manipulation (subsetting and aggregation like calculating mean) is needed. The only thing user needs to do to activate this function is to read the netCDF file using the `xr.open_dataset()` method with the keyword argument `chunks={'time':12,'lon':-1,'lat':-1}` provided. The chunk reading approach provide the opportunity to reduce the memory usage on the local machine during the calculation, the possibility of parallelizing the processes, and side-stepping the data download limit set by the OPeNDAP server (PSL server has a 500MB limit). The dataset is loaded lazily (only metadata and coordinates) shown below.In our example, we set the size of each chunk to be 12(time)x1440(lon)x720(lat) (when setting the chunk size = -1, it will use the length of the dimension as the chunksize) which is equal to 47.46 MB of data while the entire dataset is 1.39 GB. This allows us to get data in 47.46 MB chunk per download request.
The dataset is loaded lazily (only metadata and coordinates) shown below.
ds_monCalculate the SST climatology¶
First, we need to define the period that we are going to use to calculate the climatology. Here, we picked the 2019-2020 period to calculate the climatology.
Climatology
For a more accurate and scientifically valid estimate of marine heatwaves, one should usually consider a climatology period of at least 30 years. Here we set the climatology period from 2019 to 2020 (2 years) to speed up the processing time and for demonstration only. The shorter period (less memory consumption) also makes the interactive notebook launch on this page available for the user to manipulate and play with the dataset.climo_start_yr = 2019 # determine the climatology/linear trend start year
climo_end_yr = 2020 # determine the climatology/linear trend end year
ds_mon_crop = ds_mon.where((ds_mon['time.year']>=climo_start_yr)&
(ds_mon['time.year']<=climo_end_yr),drop=True)ds_mon_cropTo calculate the SST monthly climatology, we utilize the groupby method from Xarray.
ds_mon_climo = ds_mon_crop.groupby('time.month').mean()Calculate the SST anomaly¶
After the climatology is determined, we subtract the climatology from the original data to get the anomaly.
ds_mon_anom = (ds_mon_crop.groupby('time.month')-ds_mon_climo).compute().compute()
Notice the `.compute()` method in the code above. The data of SST is only loaded chunk-by-chunk, cropped to the desired period, averaged in the group of months, and finally subtracted the climatology from the original data when we execute the `.compute()` line. All these tasks are now executed in the background with a distributed server assigning tasks to different CPUs.ds_mon_anomDetermine the SST threshold based on the anomaly¶
Based on the Jacox et al., 2022, the threshold is determined based on a three month window with the center month being the monthly threhold one need to determined (e.g. January threshold is determined by all December, January, Feburary SST anomalies). Therefore, the function below is written to perform the three months window percentile operation.
########## Functions #########
# Function to calculate the 3 month rolling Quantile
def mj_3mon_quantile(da_data, mhw_threshold=90.):
da_data_quantile = xr.DataArray(coords={'lon':da_data.lon,
'lat':da_data.lat,
'month':np.arange(1,13)},
dims = ['month','lat','lon'])
for i in range(1,13):
if i == 1:
mon_range = [12,1,2]
elif i == 12 :
mon_range = [11,12,1]
else:
mon_range = [i-1,i,i+1]
da_data_quantile[i-1,:,:] = (da_data
.where((da_data['time.month'] == mon_range[0])|
(da_data['time.month'] == mon_range[1])|
(da_data['time.month'] == mon_range[2]),drop=True)
.quantile(mhw_threshold*0.01, dim = 'time', skipna = True))
return da_data_quantile
%time da_mon_quantile = mj_3mon_quantile(ds_mon_anom.sst, mhw_threshold=90)CPU times: user 9min 57s, sys: 1.33 s, total: 9min 58s
Wall time: 9min 57s
Tip
The `%time` command is jupyter cell magic to time the one-liner cell operation. It provides a great way to find the bottleneck of your data processing steps.The determined threshold value of each grid of each month is shown below
da_mon_quantile.isel(month=0).plot(vmin=0,vmax=3)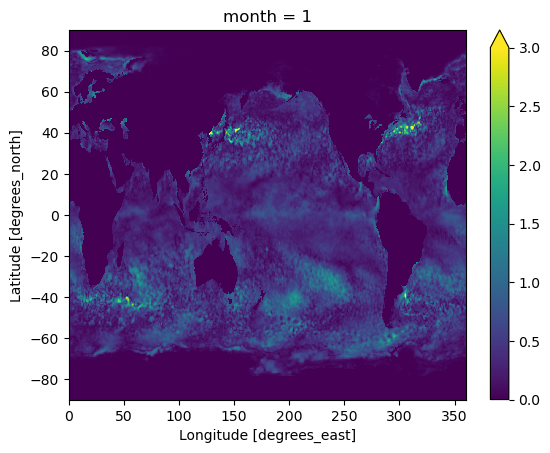
Identify the marine heatwaves based on threshold¶
The figure below shows the original SST anomaly value for the first month.
ds_mon_anom.sst.isel(time=0).plot(vmin=0,vmax=3)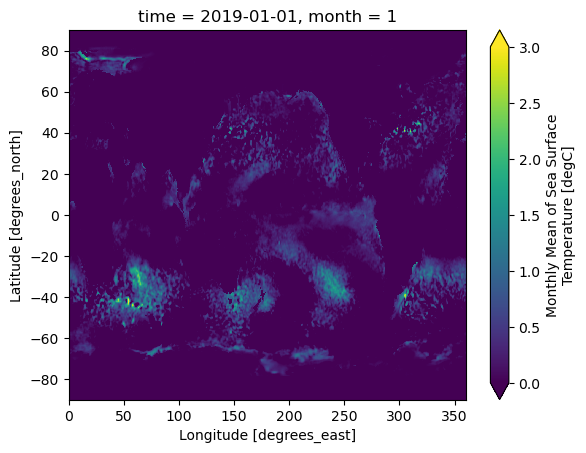
To identify the marine heatwaves based on the monthly threshold, we use the where method to find the monthly marine heatwaves with the grid that has SST anomaly below the threshold to be masked as Not-a-Number.
da_mhw = ds_mon_anom.sst.where(ds_mon_anom.sst.groupby('time.month')>da_mon_quantile)The figure below shows the SST anomalous values that are above the monthly thresholds for the first months.
da_mhw.isel(time=0).plot(vmin=0,vmax=3)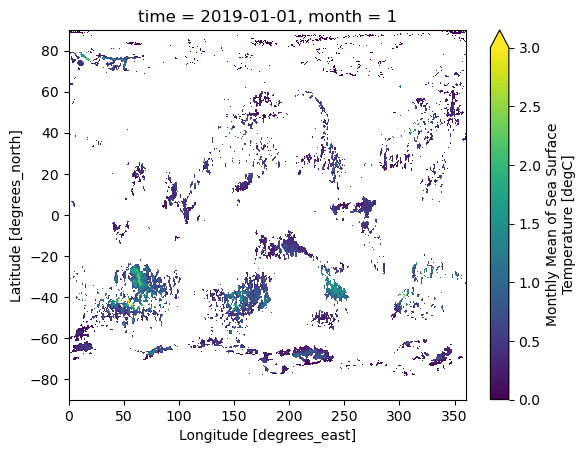
Interactive plot¶
The interactive plot is a great tool for looking at a local changes through zoom in.
Plotly provides a great interface for the user to also pin point the actual value at the point where they are interested in.
The only thing that need further data manipulation for using the plotly tool is to convert the Xarray DataArray to Pandas DataFrame.
However, this can be easily achieved throught the method .to_dataframe() provided by the Xarray package.
dff = (da_mhw.isel(time=0)
.to_dataframe()
.reset_index()
.dropna()
)
dff = dff.rename(columns={'sst':'MHW magnitude'})Plotly setting¶
After the DataFrame is created the plotly map can be created. Here, we are only using some simple options. More detail setups and options can be find on the Plotly documentation
# Setup the scatter mapbox detail
center = {'lat':38,'lon':-94} # center of the map
zoom = 2 # zoom level of the map
marker_size = 8 # marker size used on the map
mapbox_style = 'carto-positron' # mapbox options
fig = px.scatter_mapbox(dff,
lon = 'lon',
lat = 'lat',
color = 'MHW magnitude',
color_continuous_scale = 'orrd'
)
fig.update_layout(
mapbox={
'style': mapbox_style,
'center': center,
'zoom': zoom,
}
)
# Update the marker size using update_traces
fig.update_traces(marker=dict(size=marker_size))
Summary¶
Through this example, we demostrate how to lazily loaded a real world SST data from a OPeNDAP server and calculate the threshold that help us define the marine heatwave. By using the threshold, we can find the marine heatwave in each month.
What’s next?¶
A more interactive figures to view the marine heatwave will be added.
- Jacox, M. G., Alexander, M. A., Amaya, D., Becker, E., Bograd, S. J., Brodie, S., Hazen, E. L., Pozo Buil, M., & Tommasi, D. (2022). Global seasonal forecasts of marine heatwaves. Nature, 604(7906), 486–490. 10.1038/s41586-022-04573-9