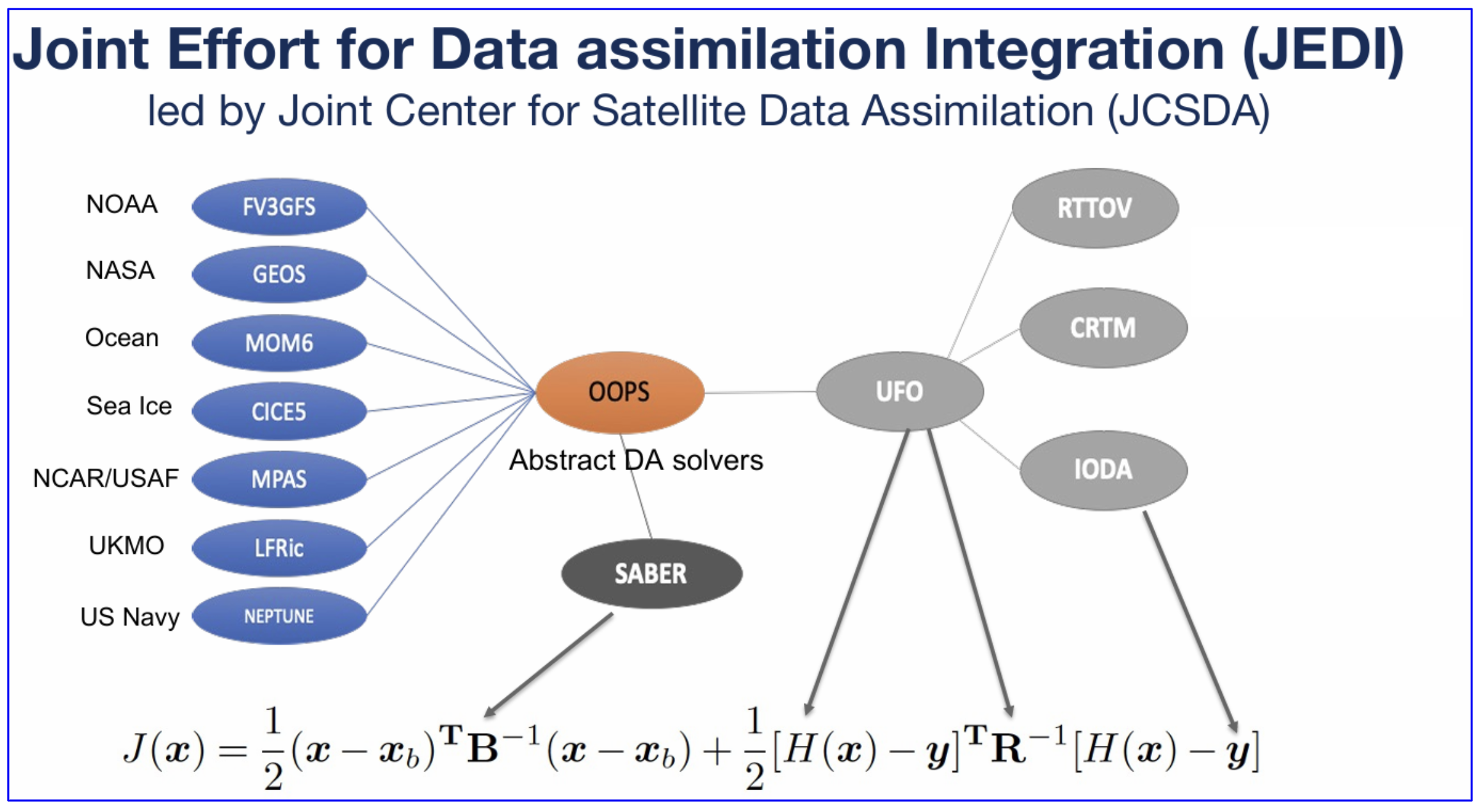
In this section, you’ll learn:¶
Utilizing UXarry to compute analysis increments, visualize increments in horizontal and vertical cross sections
Prerequisites¶
| Concepts | Importance | Notes |
|---|---|---|
| Atmospheric Data Assimilation | Helpful |
Time to learn: 10 minutes
Readers may check pyDAmonitor for more information
Import packages¶
%%time
# autoload external python modules if they changed
%load_ext autoreload
%autoreload 2
# add ../funcs to the current path
import sys, os
sys.path.append(os.path.join(os.getcwd(), ".."))
# import modules
import warnings
import math
import cartopy.crs as ccrs
import geoviews as gv
import geoviews.feature as gf
import holoviews as hv
import hvplot.xarray
from holoviews import opts
import matplotlib as mpl
import matplotlib.pyplot as plt
from matplotlib.colors import ListedColormap
import s3fs
import geopandas as gp
import numpy as np
import uxarray as ux
import xarray as xrConfigure visualization tools¶
hv.extension("bokeh")
# hv.extension("matplotlib")
# common border lines
coast_lines = gf.coastline(projection=ccrs.PlateCarree(), line_width=1, scale="50m")
state_lines = gf.states(projection=ccrs.PlateCarree(), line_width=1, line_color='gray', scale="50m")Retrieve/load MPAS/JEDI data¶
The example MPAS/JEDI data are stored at jetstream2. We need to retreive those data first.
There are two ways to retrieve MPAS data:
Download all example data from JetStream2 to local and them load them locally. This approach allows downloading the data once per machine and reuse it in notebooks.
Stream the JetStream2 S3 objects on demand. In this case, each notebook (including restarting a notebook) will retrieve the required data separately as needed.
# choose the data_load_method, check the above cell for details. Default to method 1, i.e. download once and reuse it in multiple notebooks
data_load_method = 2 # 1 or 2Method 1: Download all example data once and reuse it in mulptile notebooks¶
%%time
local_dir="/tmp"
if data_load_method == 1 and not os.path.exists(local_dir + "/conus12km/bkg/mpasout.2024-05-06_01.00.00.nc"):
jetstream_url = 'https://js2.jetstream-cloud.org:8001/'
fs = s3fs.S3FileSystem(anon=True, asynchronous=False,client_kwargs=dict(endpoint_url=jetstream_url))
conus12_path = 's3://pythia/mpas/conus12km'
fs.get(conus12_path, local_dir, recursive=True)
print("Data downloading completed")
else:
print("Skip..., either data is available in local or data_load_method is NOT 1")Skip..., either data is available in local or data_load_method is NOT 1
CPU times: user 234 μs, sys: 0 ns, total: 234 μs
Wall time: 193 μs
# Set file path
if data_load_method == 1:
grid_file = local_dir + "/conus12km/conus12km.invariant.nc_L60_GFS"
ana_file = local_dir + "/conus12km/bkg/mpasout.2024-05-06_01.00.00.nc"
bkg_file = local_dir + "/conus12km/ana/mpasout.2024-05-06_01.00.00.nc"Method 2: Stream the JetStream2 S3 objects on demand¶
%%time
if data_load_method == 2:
jetstream_url = 'https://js2.jetstream-cloud.org:8001/'
fs = s3fs.S3FileSystem(anon=True, asynchronous=False,client_kwargs=dict(endpoint_url=jetstream_url))
conus12_path = 's3://pythia/mpas/conus12km'
grid_url=f"{conus12_path}/conus12km.invariant.nc_L60_GFS"
bkg_url=f"{conus12_path}/bkg/mpasout.2024-05-06_01.00.00.nc"
ana_url=f"{conus12_path}/ana/mpasout.2024-05-06_01.00.00.nc"
grid_file = fs.open(grid_url)
ana_file = fs.open(ana_url)
bkg_file = fs.open(bkg_url)
else:
print("Skip..., data_load_method is NOT 2")CPU times: user 53.1 ms, sys: 25 ms, total: 78.1 ms
Wall time: 617 ms
Loading the data into UXarray datasets¶
We use the UXarray data structures for working with the data. This package supports data defined over unstructured grid and provides utilities for modifying and visualizing it. The available fucntionality are discussed in UxDataset documentation.
uxds_a = ux.open_dataset(grid_file, ana_file)
uxds_b = ux.open_dataset(grid_file, bkg_file)Compute the analysis increments from the JEDI data assimilation¶
JEDI updates the background atmospheric state (uxds_b) with observation innovations and gets a new atmospheric state called analysis (uxds_a).
The difference of uxds_a - uxds_b is called “analysis increments”
var_name = "theta"
uxdiff0 = uxds_a[var_name] - uxds_b[var_name]
uxvar = uxdiff0Horizontal cross sections of analysis increments at different vertical levels¶
define hcross_contour(..) and customize contour levels and color maps¶
# contour horizontal cross sections
def hcross_contour(ux_hcross, title, cmin=None, cmax=None, width=800, height=500, clevs=20, cmap="coolwarm",
symmetric_cmap=False, colorbar=True):
# Get min and max
amin = ux_hcross.min().item()
amax = ux_hcross.max().item()
title += f" min={amin:.1f} max={amax:.1f}"
cmin = math.floor(amin) if cmin is None else cmin
cmax = math.ceil(amax) if cmax is None else cmax
if symmetric_cmap: # to get a symmetric color map when cmin < 0, cmax >0
cmax = max(abs(cmin), cmax)
cmin = -cmax
if isinstance(cmap, str):
cmap = plt.get_cmap(cmap)
# generate contour plot
contour_plot = hv.operation.contours(
ux_hcross.plot(),
levels=np.linspace(cmin, cmax, num=clevs), # levels=np.arange(cmin, cmax, 0.5)
filled=True
).opts(
line_color=None, # line_width=0.001
width=width, height=height,
cmap=cmap,
clim=(cmin, cmax),
colorbar=colorbar, # cmap="inferno"
show_legend=False, tools=['hover'], title=title,
)
return contour_plot
from matplotlib.colors import ListedColormap, BoundaryNorm, to_rgba
edges = [-4, -3.5, -3, -2.5, -2, -1.5, -1, -0.5, 0, 0.5, 1.0, 1.5, 2, 2.5, 3, 3.5, 4]
colors = [
"#313695", # [-4,-3.5]
"#3f72b4", # [-3.5,-3]
"#5a9bd5", # [-3,-2.5]
"#81bfe0", # [-2.5,-2]
"#a6d8e7", # [-2,-1.5]
"#cae6ef", # [-1.5,-1]
"#e4f1f5", # [-1,-0.5]
"#f2f9fc", # [-0.5,-0.1] ← slightly pale blue
"#fcf2f2", # [0.1,0.5] ← slightly pale pink
"#f9d6d4", # [0.5,1.0]
"#f5b5b1", # [1.0,1.5]
"#ee8a85", # [1.5,2.0]
"#e75e5a", # [2.0,2.5]
"#d73027", # [2.5,3.0]
"#a50026", # [3.0,3.5]
"#67001f", # [3.5,4.0]
]
cmap = ListedColormap(colors)
plot analysis increments at different vertical levels¶
%%time
nt=0 # time dimension
plot_levels = [0, 19, 29, 39, 42, 49, 58] # [0, 29, 42] # [0, 19, 29, 39, 49, 58]
zero_shift = 0.0
plots = []
for lev in plot_levels:
dat = uxvar.isel(Time=nt, nVertLevels=lev)
tmp = hcross_contour(
dat.where((dat > 0.1) | (dat < -0.1)),
title=f'lev={lev}',
cmap=cmap,
colorbar=True,
cmax=4,
cmin=-4
)
plots.append(tmp * coast_lines * state_lines)
from IPython.display import display, Markdown
display(Markdown(
r"**Small increments** $\left[ -0.1 \ \text{to} \ 0.1 \right]$ **neglected** <br>"
r"Indicated in white spaces"
))
for p in plots:
display(p)Zoomed into Colorado using the subset capability¶
%%time
lon_center = -105.03
lat_center = 39.0
lon_incr = 5 # degree
lat_incr = 3 # degree
lon_bounds = (lon_center - lon_incr, lon_center + lon_incr)
lat_bounds = (lat_center - lat_incr, lat_center + lat_incr)
### subset to a small domain
uxdiff1 = uxdiff0.subset.bounding_box(lon_bounds, lat_bounds,)
uxvar = uxdiff1
nt=0 # time dimension
plot_levels = [0, 29, 42] # [0, 19, 29, 39, 49, 58]
plots = []
for lev in plot_levels:
tmp = hcross_contour(uxvar.isel(Time=nt, nVertLevels=lev), title=f'lev={lev}', width=700, height=500) # for the subdomain
# overlay state_lines
plots.append(tmp * coast_lines * state_lines)
# each plot has its own toolbar, which facilitates controlling each plot individually
for p in plots:
display(p)Vertical cross sections of analysis increments along an arbitary line (Great Circle Arc, GCA), a constnat laitude/longigutde¶
Along an arbitary line ( Great Circle Arc, GCA)¶
start_point = (-110, 20)
end_point = (-70, 50)
var_name = "theta"
uxdiff0 = uxds_a[var_name].isel(Time=0) - uxds_b[var_name].isel(Time=0)
uxvar = uxdiff0
cross_section_gca = uxvar.cross_section(start=start_point, end=end_point, steps=100)
hlabelticks = [
f"{abs(lat):.1f}°{'N' if lat >= 0 else 'S'}\n{abs(lon):.1f}°{'E' if lon >= 0 else 'W'}"
for lat, lon in zip(cross_section_gca['lat'], cross_section_gca['lon'])
]%matplotlib inline
fig= plt.figure(figsize=(8,3))
gs= fig.add_gridspec(1,1)
ax = fig.add_subplot(gs[0,0])
cf=ax.contourf(cross_section_gca.transpose(),cmap='coolwarm',extend='both')
tick_stride = 10
ax.set_xticks(cross_section_gca['steps'][::tick_stride])
ax.set_xticklabels(hlabelticks[::tick_stride])[Text(0, 0, '20.0°N\n110.0°W'),
Text(10, 0, '23.5°N\n107.1°W'),
Text(20, 0, '27.0°N\n104.1°W'),
Text(30, 0, '30.3°N\n100.9°W'),
Text(40, 0, '33.6°N\n97.4°W'),
Text(50, 0, '36.8°N\n93.7°W'),
Text(60, 0, '39.9°N\n89.7°W'),
Text(70, 0, '42.8°N\n85.3°W'),
Text(80, 0, '45.5°N\n80.5°W'),
Text(90, 0, '48.0°N\n75.2°W')]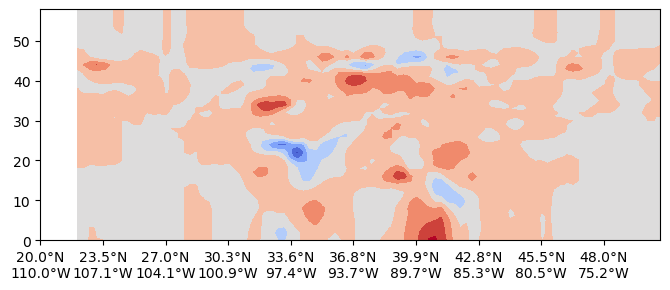
Along a constant longitude¶
lon=-83.3
cross_section = uxvar.cross_section(lon=lon, steps=100)
hlabelticks = [
f"{abs(lat):.1f}°{'N' if lat >= 0 else 'S'}" for lat in cross_section['lat']
]
%matplotlib inline
fig= plt.figure(figsize=(8,3))
gs= fig.add_gridspec(1,1)
ax = fig.add_subplot(gs[0,0])
cf=ax.contourf(cross_section.transpose(),cmap='coolwarm',extend='both')
ax.set_xticks(cross_section['steps'][::tick_stride])
ax.set_xticklabels(hlabelticks[::tick_stride])[Text(0, 0, '90.0°S'),
Text(10, 0, '71.8°S'),
Text(20, 0, '53.6°S'),
Text(30, 0, '35.5°S'),
Text(40, 0, '17.3°S'),
Text(50, 0, '0.9°N'),
Text(60, 0, '19.1°N'),
Text(70, 0, '37.3°N'),
Text(80, 0, '55.5°N'),
Text(90, 0, '73.6°N')]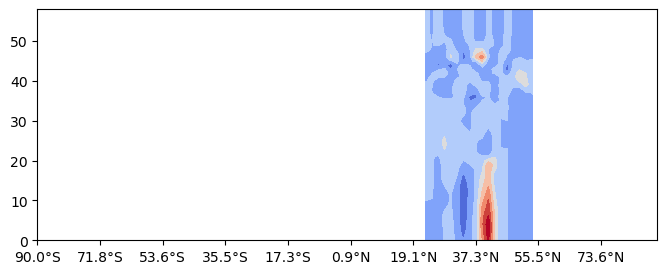
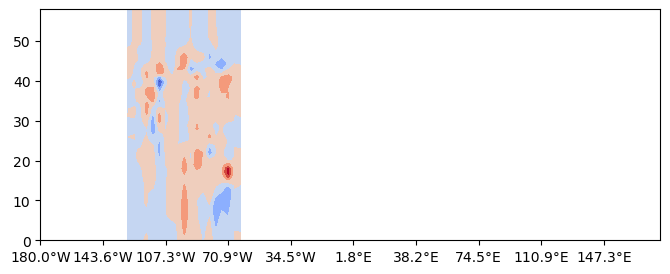
Along a constant latitude¶
lat=42.3
cross_section = uxvar.cross_section(lat=lat, steps=100)
hlabelticks = [
f"{abs(lon):.1f}°{'E' if lon >= 0 else 'W'}" for lon in cross_section['lon']
]
%matplotlib inline
fig= plt.figure(figsize=(8,3))
gs= fig.add_gridspec(1,1)
ax = fig.add_subplot(gs[0,0])
cf=ax.contourf(cross_section.transpose(),cmap='coolwarm',extend='both')
ax.set_xticks(cross_section['steps'][::tick_stride])
ax.set_xticklabels(hlabelticks[::tick_stride])[Text(0, 0, '180.0°W'),
Text(10, 0, '143.6°W'),
Text(20, 0, '107.3°W'),
Text(30, 0, '70.9°W'),
Text(40, 0, '34.5°W'),
Text(50, 0, '1.8°E'),
Text(60, 0, '38.2°E'),
Text(70, 0, '74.5°E'),
Text(80, 0, '110.9°E'),
Text(90, 0, '147.3°E')]