In this section, you’ll learn:¶
Generate data assimilation statistics and visualize them in the observation space
Prerequisites¶
| Concepts | Importance | Notes |
|---|---|---|
| Atmospheric Data Assimilation | Helpful |
Time to learn: 10 minutes
Readers may check pyDAmonitor for more information
Work with obsSpace¶
JEDI or GSI generates observation space diagnostic files, which contains original observation information, hofx (H(x), i.e. model counter-parts at the observation locations), OMB (observation minus background), OMA as well as other information.
This notebook covers how to deal with JEDI diganostic files (which are also called output ioda files). We call them jdiag files here.
Import packages¶
%%time
# autoload external python modules if they changed
%load_ext autoreload
%autoreload 2
# import modules
import warnings
import math
import sys
import os
import cartopy.crs as ccrs
import geoviews as gv
import geoviews.feature as gf
import holoviews as hv
import hvplot.xarray
from holoviews import opts
import matplotlib.pyplot as plt
import matplotlib.ticker as mticker
import matplotlib.colors as colors
from netCDF4 import Dataset
import s3fs
import seaborn as sns # seaborn handles NaN values automatically
import geopandas as gp
import numpy as np
import uxarray as ux
import xarray as xr
import pandas as pd
from obsSpace import obsSpace, fit_rate, query_data, to_dataframe, query_dataset, query_objRetrieve JEDI data¶
The example JEDI data are stored at jetstream2. We need to retreive those data first.
%%time
local_dir="/tmp/conus12km"
os.makedirs(local_dir, exist_ok=True)
if not os.path.exists(local_dir + "/jdiag_cris-fsr_n20.nc"):
jetstream_url = 'https://js2.jetstream-cloud.org:8001/'
fs = s3fs.S3FileSystem(anon=True, asynchronous=False,client_kwargs=dict(endpoint_url=jetstream_url))
conus12_path = 's3://pythia/mpas/conus12km'
fs.get(conus12_path + "/jdiag_aircar_t133.nc", local_dir+"/jdiag_aircar_t133.nc")
fs.get(conus12_path + "/jdiag_aircar_q133.nc", local_dir+"/jdiag_aircar_q133.nc")
fs.get(conus12_path + "/jdiag_aircar_uv233.nc", local_dir+"/jdiag_aircar_uv233.nc")
fs.get(conus12_path + "/jdiag_cris-fsr_n20.nc", local_dir+"/jdiag_cris-fsr_n20.nc")
print("Data downloading completed")
else:
print("Skip..., data is available in local")
jdiag_file=local_dir+"/jdiag_aircar_t133.nc"Data downloading completed
CPU times: user 857 ms, sys: 732 ms, total: 1.59 s
Wall time: 9.96 s
Use NetCDF4 to read jdiag files¶
dataset=Dataset(jdiag_file, mode='r')
query_dataset(dataset)EffectiveError0
airTemperature,
EffectiveError1
airTemperature,
EffectiveError2
airTemperature,
EffectiveQC0
airTemperature,
EffectiveQC1
airTemperature,
EffectiveQC2
airTemperature,
MetaData
stationIdentification, aircraftFlightPhase, timeOffset, prepbufrDataLvlCat, longitude, stationElevation, latitude, prepbufrReportType, pressure, longitude_latitude_pressure, dateTime, height, dumpReportType, aircraftFlightNumber,
ObsBias0
airTemperature,
ObsBias1
airTemperature,
ObsBias2
airTemperature,
ObsError
airTemperature, relativeHumidity, specificHumidity, windEastward, pressure, windNorthward,
ObsType
specificHumidity, windEastward, airTemperature, windNorthward,
ObsValue
dewpointTemperature, specificHumidity, airTemperature, windEastward, windNorthward,
QualityMarker
airTemperature, pressure, windEastward, specificHumidity, height, windNorthward,
TempObsErrorData
airTemperature,
hofx0
airTemperature,
hofx1
airTemperature,
hofx2
airTemperature,
innov1
airTemperature,
oman
airTemperature,
ombg
airTemperature,
Use the obsSpace class to read jdiag files¶
From the above output, we can see that jdiag (or output ioda file) wraps atmospheric state varaibles under attributes (or groups, such as hofx0, obsError, ObsValue, etc).
We provide a Python class obsSpace to access jdiag data in a traditional, intuitive way, such as aircar.t.hofx0, aircar.t.obsError, etc.
# create an `aircar` object using the `obsSpace` class
aircar = obsSpace(jdiag_file)check the aircar object¶
query_obj(aircar)['bt',
'dataset',
'filepath',
'groups',
'locations',
'metadata',
'nlocs',
'q',
't',
'u',
'v']From the above output, we can see the aircar object contains t, q, u, v, etc. Since jdiag_aircar_t133.nc only contains temperature observations, only aircar.t has observation diagnostics.
Query temperature data array¶
query_data(aircar.t)EffectiveError0, EffectiveError1, EffectiveError2, EffectiveQC0, EffectiveQC1, EffectiveQC2, stationIdentification, aircraftFlightPhase, timeOffset, prepbufrDataLvlCat, longitude, stationElevation, latitude, prepbufrReportType, pressure, dateTime, height, dumpReportType, aircraftFlightNumber, ObsBias0, ObsBias1, ObsBias2, ObsError, ObsType, ObsValue, QualityMarker, TempObsErrorData, hofx0, hofx1, hofx2, innov1, oman, ombg,
# print out array values
np.set_printoptions(threshold=500) # don't print out all array values
print(aircar.t.ObsValue)[229.15 220.95 219.95 ... 271.94998 270.15 232.15 ]
Convert data array to Pandas DataFrame¶
Converting jdiag data into Pandas DataFrame brings up lots of benefits and we can utilize lots of existing DataFrame capabilities.
df = to_dataframe(aircar.t)
dfPlot histograms¶
Example 1¶
# plot histogram of OmA
plt.figure(figsize=(8, 5))
#sns.histplot(df["oman"], bins=50, kde=True, color="steelblue")
sns.histplot(aircar.t.oman, bins=100, kde=True, color="steelblue")
plt.title("Histogram of oman")
plt.xlabel("oman values")
plt.ylabel("Density")
plt.tight_layout()
plt.show()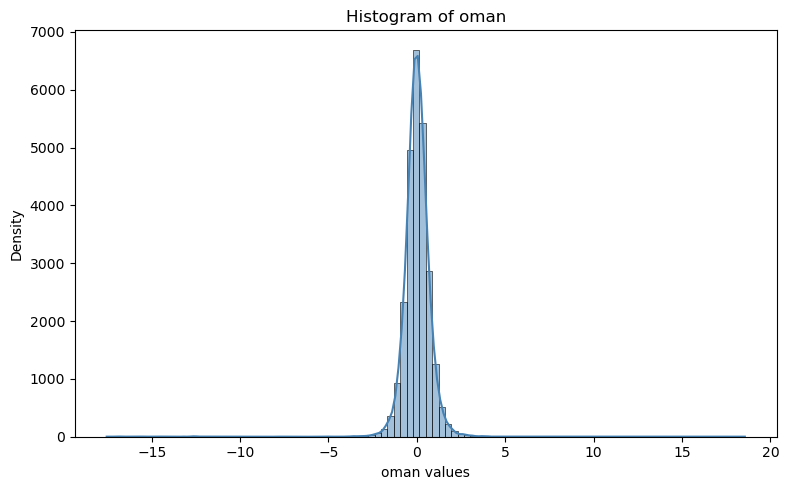
Example 2¶
# overlay OMB and OMA histogram together
df_long = df[["oman", "ombg"]].melt(var_name="variable", value_name="value")
plt.figure(figsize=(8, 5))
sns.histplot(data=df_long, x="value", hue="variable", bins=50, kde=True, element="step", stat="count")
plt.title("Overlayed Histogram: oman vs ombg")
plt.tight_layout()
plt.show()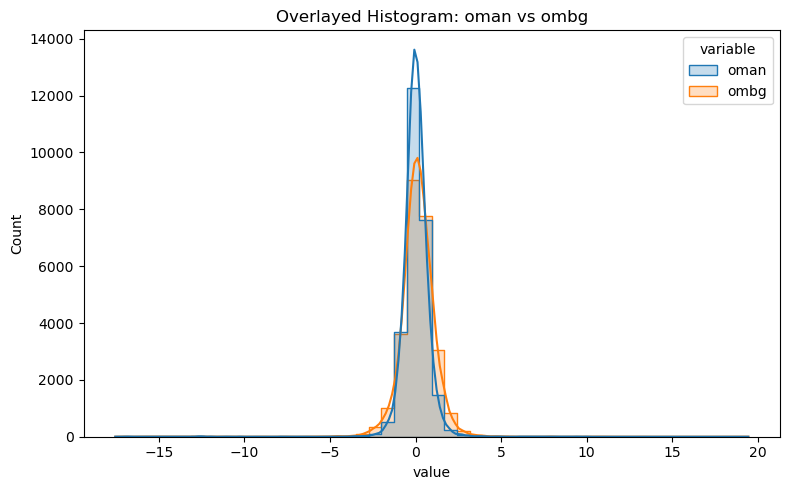
Example 3¶
# overlay OMB and OMA histogram together
plt.figure(figsize=(8, 5))
sns.histplot(df["oman"], bins=100, kde=True, color="blue", label="oman", multiple="layer")
sns.histplot(df["ombg"], bins=100, kde=True, color="red", label="ombg", multiple="layer")
plt.title("Overlayed Histogram: oman vs ombg")
plt.xlabel("Value")
plt.ylabel("Frequency")
plt.legend()
plt.tight_layout()
plt.show()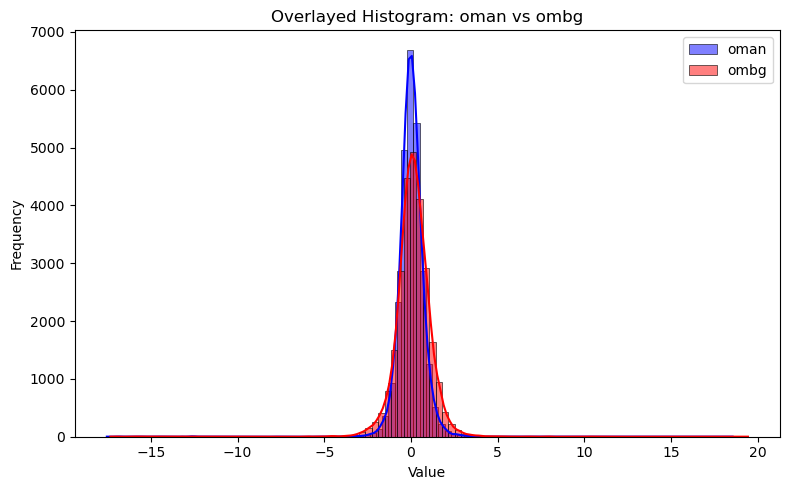
plot fitting rate¶
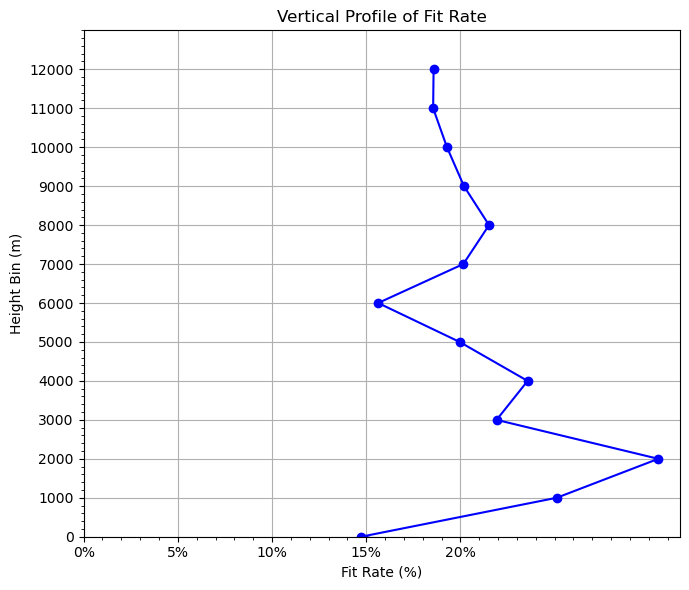
The rate fitting to observations is an important metric to evaluate data assimilation performance. We don’t want a small fitting rate, which means very little observation impacts on the model forecast. We don’t want a large fitting rate either, which means we fit too close to the observations and the model forecast will crash. Usually, a fitting rate of 20%~30% is expected.
# 1. Filter valid data (both 'oman' and 'ombg' are not NaN)
valid_df = df[df["oman"].notna() & df["ombg"].notna()].copy()
valid_df = valid_df.dropna(subset=["height"]) # removes any rows in valid_df where height is missing (NaN)
print(valid_df[valid_df["height"] < 0]["height"]) # negative height19 -57
140 -27
141 -6
196 -15
325 -34
..
23364 -58
23854 -48
24839 -24
25899 -34
26007 -32
Name: height, Length: 69, dtype: int32
dz = 1000
grouped = fit_rate(aircar.t, dz=dz)
# 5. Plot vertical profile of fit_rate vs height
plt.figure(figsize=(7, 6))
plt.plot(grouped["fit_rate"], grouped["height_bin"], marker="o", color="blue")
# plt.axvline(x=0, color="gray", linestyle="--") # ax vertical line
plt.xlabel("Fit Rate (%)") # label change
plt.gca().xaxis.set_major_formatter(plt.FuncFormatter(lambda x, _: f'{x*100:.0f}%')) # format as %
plt.ylabel("Height Bin (m)")
plt.title("Vertical Profile of Fit Rate")
# Fine-tune ticks
plt.xticks(np.arange(0, 0.25, 0.05)) #, fontsize=12)
plt.yticks(np.arange(0, 13000, dz)) #, , fontsize=12)
# Add minor ticks
from matplotlib.ticker import AutoMinorLocator
plt.gca().xaxis.set_minor_locator(AutoMinorLocator())
plt.gca().yaxis.set_minor_locator(AutoMinorLocator())
# plt.grid(which='both', linestyle='--', linewidth=0.5)
plt.grid(True)
plt.ylim(0, 13000) # set y-axis from 0 (bottom) to 13,000 (top)
plt.tight_layout()
plt.show()OMA: bias=0.0063 rms=0.9397
OMB: bias=0.1196 rms=1.1644
Overall fit_rate: 19.3006%
0, -1000, 1.0326, 1.4933, 30.8519%
1, 0, 1.5019, 1.7613, 14.7300%
2, 1000, 0.8644, 1.1543, 25.1132%
3, 2000, 0.7517, 1.0818, 30.5067%
4, 3000, 0.7249, 0.9284, 21.9214%
5, 4000, 0.7171, 0.9383, 23.5695%
6, 5000, 1.0533, 1.3160, 19.9605%
7, 6000, 1.1809, 1.3997, 15.6337%
8, 7000, 0.7965, 0.9975, 20.1548%
9, 8000, 0.5794, 0.7382, 21.5208%
10, 9000, 0.6157, 0.7714, 20.1891%
11, 10000, 0.6812, 0.8440, 19.2867%
12, 11000, 0.7952, 0.9764, 18.5534%
13, 12000, 1.2666, 1.5556, 18.5768%
print(grouped["height_bin"])0 -1000
1 0
2 1000
3 2000
4 3000
5 4000
6 5000
7 6000
8 7000
9 8000
10 9000
11 10000
12 11000
13 12000
Name: height_bin, dtype: int32
Plot satellite radiance observations¶
load satellite data using obsSpace¶
%%time
obsCris = obsSpace(local_dir + "/jdiag_cris-fsr_n20.nc")CPU times: user 3.23 s, sys: 225 ms, total: 3.45 s
Wall time: 3.45 s
# query the `bt` data array, excluding the metadata starting with `sensorCentralWavenumber_`. bt=Brightness Temperature
query_data(obsCris.bt, meta_exclude="sensorCentralWavenumber_")DerivedObsError, DerivedObsValue, CloudDetectMinResidualIR, GeoDomainCheck, NearSSTRetCheckIR, ScanEdgeRemoval, SurfaceTempJacobianCheck, GrossCheck, Thinning, UseflagCheck, EffectiveError0, EffectiveError1, EffectiveError2, EffectiveQC0, EffectiveQC1, EffectiveQC2, cloudCoverTotal, sensorZenithAngle, satelliteIdentifier, sensorScanPosition, heightOfTopOfCloud, instrumentIdentifier, longitude, latitude, sensorViewAngle, scanLineNumber, solarZenithAngle, fieldOfRegardNumber, sensorChannelNumber, sensorAzimuthAngle, fieldOfViewNumber, dateTime, solarAzimuthAngle, ObsBias0, ObsBias1, ObsBias2, constantPredictor, emissivityJacobianPredictor, hofx0, hofx1, hofx2, innov1, lapseRatePredictor, lapseRate_order_2Predictor, oman, ombg, sensorScanAnglePredictor, sensorScanAngle_order_2Predictor, sensorScanAngle_order_3Predictor, sensorScanAngle_order_4Predictor,
# print H(x) values
print(obsCris.bt.hofx0)[[-- -- -- ... -- -- --]
[-- -- -- ... -- -- --]
[-- -- -- ... -- -- --]
...
[225.10011291503906 228.2823944091797 226.98941040039062 ...
275.4639587402344 275.5229797363281 275.2835998535156]
[225.03855895996094 228.01905822753906 226.7443084716797 ...
285.9146423339844 285.9745788574219 285.7776794433594]
[223.8397216796875 226.27716064453125 224.85659790039062 ...
285.6235656738281 285.6897277832031 285.5458679199219]]
assemble target obs array¶
ncount=0
idx = []
idx2 = []
ch=61
for n in np.arange(len(obsCris.bt.ombg[:,ch])):
#if obsCris.bt.CloudDetectMinResidualIR[n,ch] == 1:
if obsCris.bt.ombg[n,ch] > -200 and obsCris.bt.ombg[n,ch] < 200:
idx.append(n)
ncount = ncount + 1
lat=obsCris.bt.latitude[idx]
lon=obsCris.bt.longitude[idx]
obarray=obsCris.bt.DerivedObsValue[idx,ch]
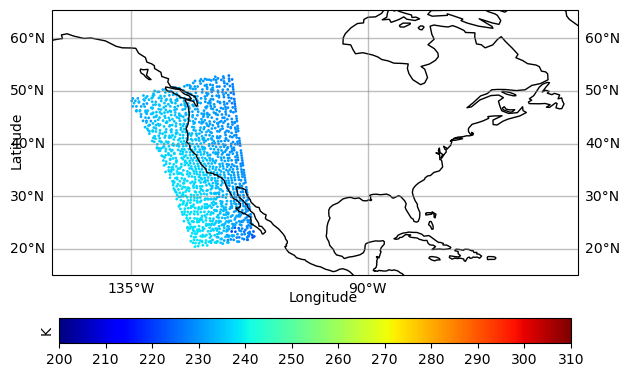
print(lon,lat,obarray)
print(ncount)[-115.9189 -119.51454 -123.75143 ... -120.8286 -122.65434 -132.61992] [39.96 40.04689 23.87319 ... 48.48392 49.66428 49.14518] [229.50182 232.08597 237.9213 ... 230.9885 231.4381 233.62317]
1313
prepare color map¶
datmi = np.nanmin(obarray) # Min of the data
datma = np.nanmax(obarray) # Max of the data
import matplotlib.pyplot as plt
if np.nanmin(obarray) < 0:
cmax = datma
cmin = datmi
cmax=310
cmin=200
#cmax=1.0
#cmin=-1.0
cmap = 'RdBu'
else:
#cmax = omean+stdev
#cmin = np.maximum(omean-stdev, 0.0)
cma = datma
cmin = datmi
cmax=310
cmin=200
#cmax=1.0
#cmin=-1.0
cmap = 'RdBu'
cmap = 'viridis'
cmap = 'jet'
cmin = 200.
cmax = 310.
conus_12km = [-150, -50, 15, 55]
color_map = plt.cm.get_cmap(cmap)
reversed_color_map = color_map.reversed()
units = 'K'
#units = '%'
fig = plt.figure(figsize=(10, 5))/tmp/ipykernel_3932/2108599323.py:33: MatplotlibDeprecationWarning: The get_cmap function was deprecated in Matplotlib 3.7 and will be removed in 3.11. Use ``matplotlib.colormaps[name]`` or ``matplotlib.colormaps.get_cmap()`` or ``pyplot.get_cmap()`` instead.
color_map = plt.cm.get_cmap(cmap)
<Figure size 1000x500 with 0 Axes>Make the plot¶
# Initialize the plot pointing to the projection
# ------------------------------------------------
ax = plt.axes(projection=ccrs.PlateCarree(central_longitude=0))
# Plot grid lines
# ----------------
gl = ax.gridlines(crs=ccrs.PlateCarree(central_longitude=0), draw_labels=True,
linewidth=1, color='gray', alpha=0.5, linestyle='-')
gl.top_labels = False
gl.xlabel_style = {'size': 10, 'color': 'black'}
gl.ylabel_style = {'size': 10, 'color': 'black'}
gl.xlocator = mticker.FixedLocator(
[-180, -135, -90, -45, 0, 45, 90, 135, 179.9])
ax.set_ylabel("Latitude", fontsize=7)
ax.set_xlabel("Longitude", fontsize=7)
# Get scatter data
# ------------------
#print('obarray = ', obarray)
print('min/max obarray = ', min(obarray),max(obarray))
#sc = ax.scatter(lonData, latData,
sc = ax.scatter(lon, lat,
c=obarray, s=4, linewidth=0,
transform=ccrs.PlateCarree(), cmap=cmap, vmin=cmin, vmax = cmax, norm=None, antialiased=True)
# Plot colorbar
# --------------
cbar = plt.colorbar(sc, ax=ax, orientation="horizontal", pad=.1, fraction=0.06,ticks=[200, 210, 220, 230, 240, 250, 260, 270, 280, 290, 300, 310])
#cbar = plt.colorbar(sc, ax=ax, orientation="horizontal", pad=.1, fraction=0.06,ticks=[-3, -2.5, -2, -1.5, -1, -0.5, 0, 0.5, 1.0, 1.5, 2.0, 2.5, 3 ])
#cbar = plt.colorbar(sc, ax=ax, orientation="horizontal", pad=.1, fraction=0.06,ticks=[0, 10, 20, 20, 40, 50, 60, 70, 80, 90, 100])
cbar.ax.set_ylabel(units, fontsize=10)
# Plot globally
# --------------
#ax.set_global()
#ax.set_extent(conus)
ax.set_extent(conus_12km)
# Draw coastlines
# ----------------
ax.coastlines()
ax.text(0.45, -0.1, 'Longitude', transform=ax.transAxes, ha='left')
ax.text(-0.08, 0.4, 'Latitude', transform=ax.transAxes,
rotation='vertical', va='bottom')
#text = f"Total Count:{datcont:0.0f}, Max/Min/Mean/Std: {datma:0.3f}/{datmi:0.3f}/{omean:0.3f}/{stdev:0.3f} {units}"
#print(text)
#ax.text(0.67, -0.1, text, transform=ax.transAxes, va='bottom', fontsize=6.2)
dpi=150
gl.top_labels = False
plt.tight_layout()
# show plot
# -----------
# pname='test.png'
# plt.savefig(pname, dpi=dpi) min/max obarray = 220.04314 238.75264
/home/runner/micromamba/envs/mpas-jedi-cookbook-dev/lib/python3.11/site-packages/cartopy/io/__init__.py:242: DownloadWarning: Downloading: https://naturalearth.s3.amazonaws.com/110m_physical/ne_110m_coastline.zip
warnings.warn(f'Downloading: {url}', DownloadWarning)