Explore WRF simulations of the surface albedo¶
Import modules¶
from datetime import timedelta
import cmweather
import xarray as xr
import xwrf
import glob
import matplotlib.pyplot as plt
from matplotlib import cm
import numpy as npLoad WRF data in January 2022 and April 2023¶
files_202201 = sorted(glob.glob("/data/home/yxie/wrf_data/jan_2022/*"))
files_202304 = sorted(glob.glob("/data/home/yxie/wrf_data/apr_2023/*"))# pickd dates for each event: onset and five days after that
files_case1 = files_202201[1:7] # daily
files_case2 = files_202201[24:30] # daily
files_case3 = files_202304[8:32] # every six hours# double check the dates
files_case1['/data/home/yxie/wrf_data/jan_2022/wrfhourly_d03_2022-01-02.nc',
'/data/home/yxie/wrf_data/jan_2022/wrfhourly_d03_2022-01-03.nc',
'/data/home/yxie/wrf_data/jan_2022/wrfhourly_d03_2022-01-04.nc',
'/data/home/yxie/wrf_data/jan_2022/wrfhourly_d03_2022-01-05.nc',
'/data/home/yxie/wrf_data/jan_2022/wrfhourly_d03_2022-01-06.nc',
'/data/home/yxie/wrf_data/jan_2022/wrfhourly_d03_2022-01-07.nc']files_case2['/data/home/yxie/wrf_data/jan_2022/wrfhourly_d03_2022-01-25.nc',
'/data/home/yxie/wrf_data/jan_2022/wrfhourly_d03_2022-01-26.nc',
'/data/home/yxie/wrf_data/jan_2022/wrfhourly_d03_2022-01-27.nc',
'/data/home/yxie/wrf_data/jan_2022/wrfhourly_d03_2022-01-28.nc',
'/data/home/yxie/wrf_data/jan_2022/wrfhourly_d03_2022-01-29.nc',
'/data/home/yxie/wrf_data/jan_2022/wrfhourly_d03_2022-01-30.nc']files_case3['/data/home/yxie/wrf_data/apr_2023/wrfout_d03_2023-04-03_00:00:00',
'/data/home/yxie/wrf_data/apr_2023/wrfout_d03_2023-04-03_06:00:00',
'/data/home/yxie/wrf_data/apr_2023/wrfout_d03_2023-04-03_12:00:00',
'/data/home/yxie/wrf_data/apr_2023/wrfout_d03_2023-04-03_18:00:00',
'/data/home/yxie/wrf_data/apr_2023/wrfout_d03_2023-04-04_00:00:00',
'/data/home/yxie/wrf_data/apr_2023/wrfout_d03_2023-04-04_06:00:00',
'/data/home/yxie/wrf_data/apr_2023/wrfout_d03_2023-04-04_12:00:00',
'/data/home/yxie/wrf_data/apr_2023/wrfout_d03_2023-04-04_18:00:00',
'/data/home/yxie/wrf_data/apr_2023/wrfout_d03_2023-04-05_00:00:00',
'/data/home/yxie/wrf_data/apr_2023/wrfout_d03_2023-04-05_06:00:00',
'/data/home/yxie/wrf_data/apr_2023/wrfout_d03_2023-04-05_12:00:00',
'/data/home/yxie/wrf_data/apr_2023/wrfout_d03_2023-04-05_18:00:00',
'/data/home/yxie/wrf_data/apr_2023/wrfout_d03_2023-04-06_00:00:00',
'/data/home/yxie/wrf_data/apr_2023/wrfout_d03_2023-04-06_06:00:00',
'/data/home/yxie/wrf_data/apr_2023/wrfout_d03_2023-04-06_12:00:00',
'/data/home/yxie/wrf_data/apr_2023/wrfout_d03_2023-04-06_18:00:00',
'/data/home/yxie/wrf_data/apr_2023/wrfout_d03_2023-04-07_00:00:00',
'/data/home/yxie/wrf_data/apr_2023/wrfout_d03_2023-04-07_06:00:00',
'/data/home/yxie/wrf_data/apr_2023/wrfout_d03_2023-04-07_12:00:00',
'/data/home/yxie/wrf_data/apr_2023/wrfout_d03_2023-04-07_18:00:00',
'/data/home/yxie/wrf_data/apr_2023/wrfout_d03_2023-04-08_00:00:00',
'/data/home/yxie/wrf_data/apr_2023/wrfout_d03_2023-04-08_06:00:00',
'/data/home/yxie/wrf_data/apr_2023/wrfout_d03_2023-04-08_12:00:00',
'/data/home/yxie/wrf_data/apr_2023/wrfout_d03_2023-04-08_18:00:00']ds = xr.open_dataset(files_case1[0]).xwrf.postprocess()
#ds = xr.open_dataset(files_case1[0])
dsLoading...
Visualize the snow albedo data¶
ds.ALBEDOLoading...
Visualize snow albedo for the three cases¶
Case 1: control event - Jan 2-8, 2022
Case 2: black carbon event - Jan 25-31, 2022
Case 3: dust event - Apr 3-9, 2023
dscase1_ini = xr.open_dataset(files_case1[0]).xwrf.postprocess()
dscase1_end = xr.open_dataset(files_case1[-1]).xwrf.postprocess()
dscase2_ini = xr.open_dataset(files_case2[0]).xwrf.postprocess()
dscase2_end = xr.open_dataset(files_case2[-1]).xwrf.postprocess()
# choose 18 PM UTC, which corresponds to 12 PM in local time
dscase3_ini = xr.open_dataset(files_case3[3]).xwrf.postprocess()
dscase3_end = xr.open_dataset(files_case3[-1]).xwrf.postprocess()Time series of simulated snow albedo¶
Case 1: Control event
for itt in range(0, len(files_case1) ):
ds = xr.open_dataset(files_case1[itt]).xwrf.postprocess()
alb = ds.ALBEDO[:,124, 109].squeeze().values
time = ds.Time.squeeze().values
if itt == 0:
alb1 = alb
time1 = time
else:
alb1 = np.append(alb1, alb)
time1 = np.append(time1, time)
Visualize the time series
plt.figure(figsize=(5.5,2),dpi=150)
plt.plot(time1, alb1,'.-')
plt.xticks(fontsize=7)
plt.yticks(np.arange(0.7,0.9,0.05),fontsize=8)
plt.ylabel('WRF albedo')
plt.grid()
plt.title('Control event')
plt.show()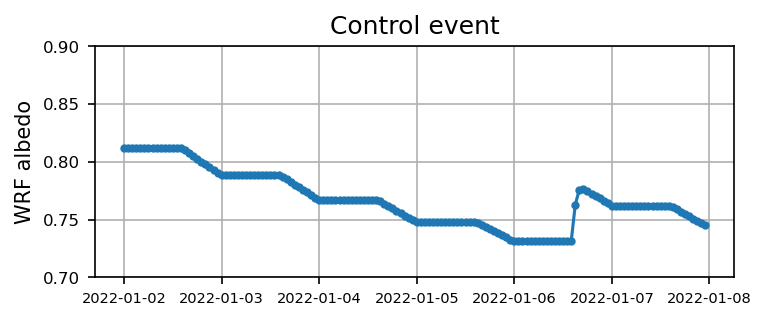
Case 2: Black carbon event
for itt in range(0, len(files_case2) ):
ds = xr.open_dataset(files_case2[itt]).xwrf.postprocess()
alb = ds.ALBEDO[:,124,109].squeeze().values
time = ds.Time.squeeze().values
if itt == 0:
alb2 = alb
time2 = time
else:
alb2 = np.append(alb2, alb)
time2 = np.append(time2, time)
plt.figure(figsize=(5.5,2),dpi=150)
plt.plot(time2, alb2,'.-')
plt.xticks(fontsize=7)
plt.yticks(np.arange(0.6,0.8,0.05),fontsize=8)
plt.ylabel('WRF albedo')
plt.grid()
plt.title('Black carbon event')
plt.show()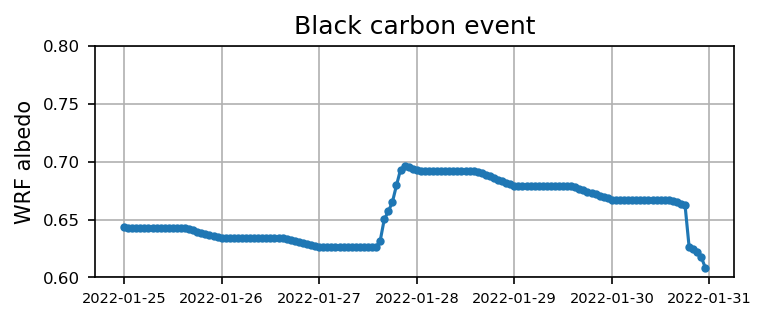
Case 3: Dust event
for itt in range(0, len(files_case3) ):
ds = xr.open_dataset(files_case3[itt]).xwrf.postprocess()
alb = ds.ALBEDO[:,124,109].squeeze().values
time = ds.Time.squeeze().values
if itt == 0:
alb3 = alb
time3 = time
else:
alb3 = np.append(alb3, alb)
time3 = np.append(time3, time)
plt.figure(figsize=(5.5,2),dpi=150)
plt.plot(time3, alb3,'.-')
plt.xticks(fontsize=7)
plt.yticks(np.arange(0.7,0.9,0.05),fontsize=8)
plt.ylabel('WRF albedo')
plt.grid()
plt.title('Dust event')
plt.show()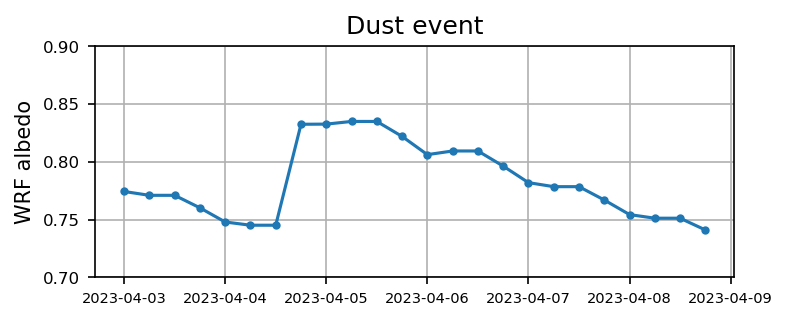
Visualize albedo time series combination¶
plt.figure(figsize=(5,2), dpi=150)
plt.plot(np.arange(0, alb1.size)/(alb1.size-1)*5 + 1, alb1, '--', linewidth=1, label='control event')
plt.plot(np.arange(0, alb2.size)/(alb2.size-1)*5 + 1, alb2, 'o-', markersize=2, label='black carbon event')
plt.plot(np.arange(0, alb3.size)/(alb3.size-1)*5 + 1, alb3, '^-', markersize=3, label='dust event')
plt.xlabel('days')
plt.ylabel('WRF albedo')
plt.legend(fontsize=8)
plt.yticks(np.arange(0.65, 1.00, 0.05))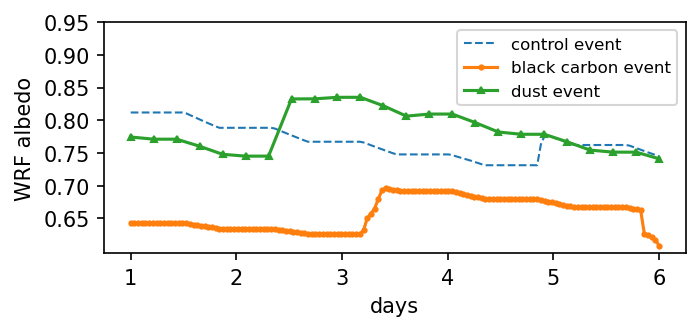
# get the longitude and latitude coordinates
xlong = dscase3_ini.XLONG.copy()
xlat = dscase3_ini.XLAT.copy()xlong
xlatLoading...
xlong.shape(201, 201)xlat[124,109]Loading...
xlong[124,109]Loading...
dscase1_ini.ALBEDO.mean(dim=["Time"])Loading...
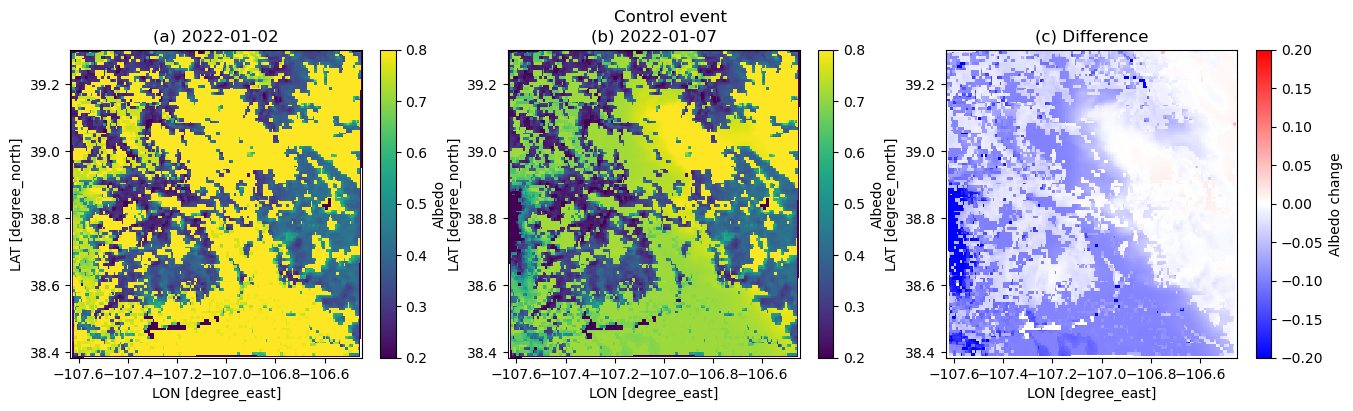
plt.figure(figsize=(16,4))
albini = dscase1_ini.ALBEDO[17,:,:]
albend = dscase1_end.ALBEDO[17,:,:]
plt.suptitle('Control event')
plt.subplot(1,3,1)
plt.pcolormesh(xlong, xlat, albini.squeeze(), vmin=0.2, vmax=0.8)
plt.xlabel('LON [degree_east]', fontsize=10)
plt.ylabel('LAT [degree_north]', fontsize=10)
plt.colorbar(label='Albedo')
plt.title('(a) 2022-01-02')
plt.subplot(1,3,2)
plt.pcolormesh(xlong, xlat, albend.squeeze(), vmin=0.2, vmax=0.8)
plt.xlabel('LON [degree_east]', fontsize=10)
plt.ylabel('LAT [degree_north]', fontsize=10)
plt.colorbar(label='Albedo')
plt.title('(b) 2022-01-07')
plt.subplot(1,3,3)
plt.pcolormesh(xlong, xlat, albend.squeeze() - albini.squeeze(), cmap=cm.bwr, vmin=-0.2, vmax=0.2)
plt.xlabel('LON [degree_east]', fontsize=10)
plt.ylabel('LAT [degree_north]', fontsize=10)
plt.colorbar(label='Albedo change')
plt.title('(c) Difference')/tmp/ipykernel_3207/648137882.py:8: UserWarning: The input coordinates to pcolormesh are interpreted as cell centers, but are not monotonically increasing or decreasing. This may lead to incorrectly calculated cell edges, in which case, please supply explicit cell edges to pcolormesh.
plt.pcolormesh(xlong, xlat, albini.squeeze(), vmin=0.2, vmax=0.8)
/tmp/ipykernel_3207/648137882.py:15: UserWarning: The input coordinates to pcolormesh are interpreted as cell centers, but are not monotonically increasing or decreasing. This may lead to incorrectly calculated cell edges, in which case, please supply explicit cell edges to pcolormesh.
plt.pcolormesh(xlong, xlat, albend.squeeze(), vmin=0.2, vmax=0.8)
/tmp/ipykernel_3207/648137882.py:22: UserWarning: The input coordinates to pcolormesh are interpreted as cell centers, but are not monotonically increasing or decreasing. This may lead to incorrectly calculated cell edges, in which case, please supply explicit cell edges to pcolormesh.
plt.pcolormesh(xlong, xlat, albend.squeeze() - albini.squeeze(), cmap=cm.bwr, vmin=-0.2, vmax=0.2)
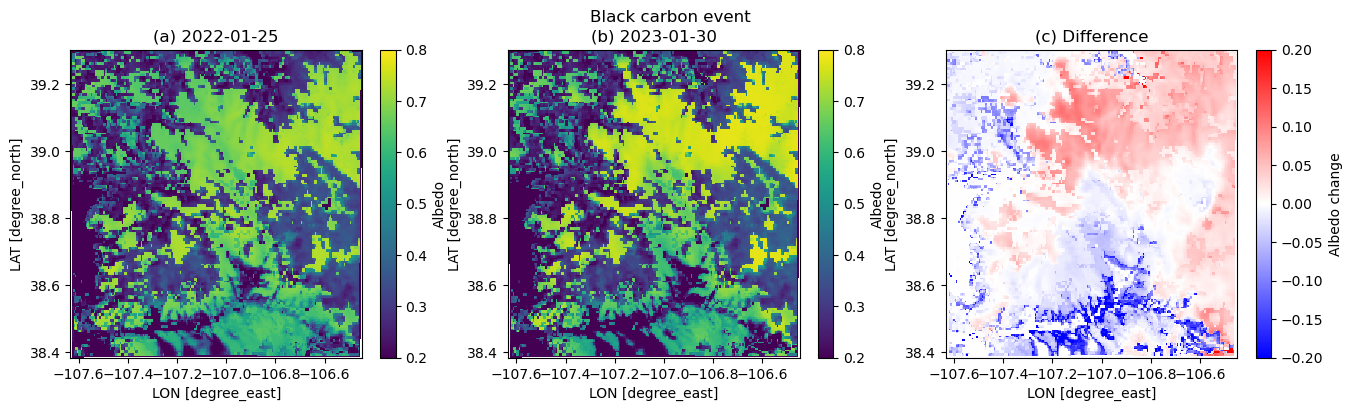
plt.figure(figsize=(16,4))
albini = dscase2_ini.ALBEDO[17,:,:]
albend = dscase2_end.ALBEDO[17,:,:]
plt.suptitle('Black carbon event')
plt.subplot(1,3,1)
plt.pcolormesh(xlong, xlat, albini.squeeze(), vmin=0.2, vmax=0.8)
plt.xlabel('LON [degree_east]', fontsize=10)
plt.ylabel('LAT [degree_north]', fontsize=10)
plt.colorbar(label='Albedo')
plt.title('(a) 2022-01-25')
plt.subplot(1,3,2)
plt.pcolormesh(xlong, xlat, albend.squeeze(), vmin=0.2, vmax=0.8)
plt.xlabel('LON [degree_east]', fontsize=10)
plt.ylabel('LAT [degree_north]', fontsize=10)
plt.colorbar(label='Albedo')
plt.title('(b) 2023-01-30')
plt.subplot(1,3,3)
plt.pcolormesh(xlong, xlat, albend.squeeze() - albini.squeeze(), cmap=cm.bwr, vmin=-0.2, vmax=0.2)
plt.xlabel('LON [degree_east]', fontsize=10)
plt.ylabel('LAT [degree_north]', fontsize=10)
plt.colorbar(label='Albedo change')
plt.title('(c) Difference')/tmp/ipykernel_3207/78071660.py:8: UserWarning: The input coordinates to pcolormesh are interpreted as cell centers, but are not monotonically increasing or decreasing. This may lead to incorrectly calculated cell edges, in which case, please supply explicit cell edges to pcolormesh.
plt.pcolormesh(xlong, xlat, albini.squeeze(), vmin=0.2, vmax=0.8)
/tmp/ipykernel_3207/78071660.py:15: UserWarning: The input coordinates to pcolormesh are interpreted as cell centers, but are not monotonically increasing or decreasing. This may lead to incorrectly calculated cell edges, in which case, please supply explicit cell edges to pcolormesh.
plt.pcolormesh(xlong, xlat, albend.squeeze(), vmin=0.2, vmax=0.8)
/tmp/ipykernel_3207/78071660.py:22: UserWarning: The input coordinates to pcolormesh are interpreted as cell centers, but are not monotonically increasing or decreasing. This may lead to incorrectly calculated cell edges, in which case, please supply explicit cell edges to pcolormesh.
plt.pcolormesh(xlong, xlat, albend.squeeze() - albini.squeeze(), cmap=cm.bwr, vmin=-0.2, vmax=0.2)
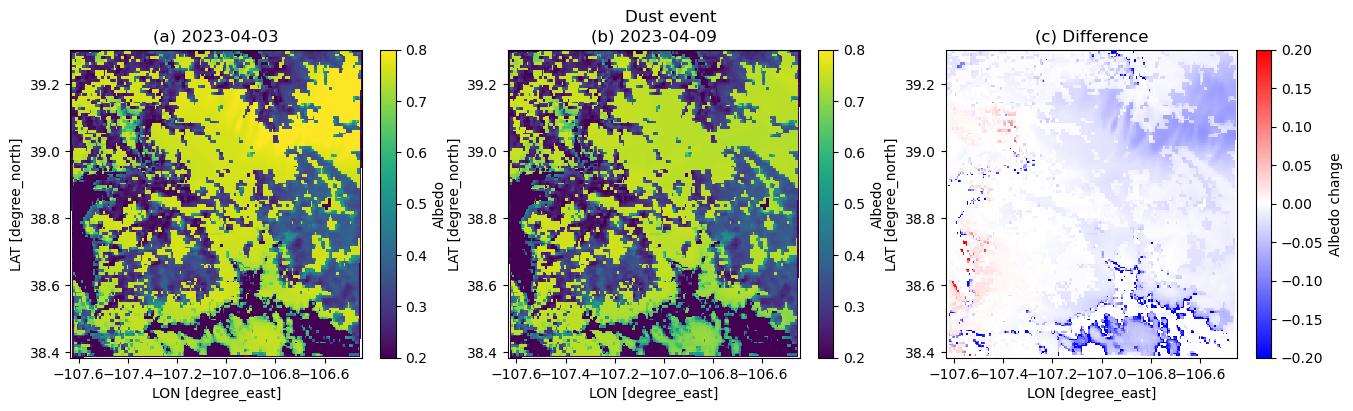
plt.figure(figsize=(16,4))
plt.suptitle('Dust event')
plt.subplot(1,3,1)
plt.pcolormesh(xlong, xlat, dscase3_ini.ALBEDO.squeeze(), vmin=0.2, vmax=0.8)
plt.xlabel('LON [degree_east]', fontsize=10)
plt.ylabel('LAT [degree_north]', fontsize=10)
plt.colorbar(label='Albedo')
plt.title('(a) 2023-04-03')
plt.subplot(1,3,2)
plt.pcolormesh(xlong, xlat, dscase3_end.ALBEDO.squeeze(), vmin=0.2, vmax=0.8)
plt.xlabel('LON [degree_east]', fontsize=10)
plt.ylabel('LAT [degree_north]', fontsize=10)
plt.colorbar(label='Albedo')
plt.title('(b) 2023-04-09')
plt.subplot(1,3,3)
plt.pcolormesh(xlong, xlat, dscase3_end.ALBEDO.squeeze() - dscase3_ini.ALBEDO.squeeze(), cmap=cm.bwr, vmin=-0.2, vmax=0.2)
plt.xlabel('LON [degree_east]', fontsize=10)
plt.ylabel('LAT [degree_north]', fontsize=10)
plt.colorbar(label='Albedo change')
plt.title('(c) Difference')/tmp/ipykernel_3207/964711993.py:6: UserWarning: The input coordinates to pcolormesh are interpreted as cell centers, but are not monotonically increasing or decreasing. This may lead to incorrectly calculated cell edges, in which case, please supply explicit cell edges to pcolormesh.
plt.pcolormesh(xlong, xlat, dscase3_ini.ALBEDO.squeeze(), vmin=0.2, vmax=0.8)
/tmp/ipykernel_3207/964711993.py:13: UserWarning: The input coordinates to pcolormesh are interpreted as cell centers, but are not monotonically increasing or decreasing. This may lead to incorrectly calculated cell edges, in which case, please supply explicit cell edges to pcolormesh.
plt.pcolormesh(xlong, xlat, dscase3_end.ALBEDO.squeeze(), vmin=0.2, vmax=0.8)
/tmp/ipykernel_3207/964711993.py:20: UserWarning: The input coordinates to pcolormesh are interpreted as cell centers, but are not monotonically increasing or decreasing. This may lead to incorrectly calculated cell edges, in which case, please supply explicit cell edges to pcolormesh.
plt.pcolormesh(xlong, xlat, dscase3_end.ALBEDO.squeeze() - dscase3_ini.ALBEDO.squeeze(), cmap=cm.bwr, vmin=-0.2, vmax=0.2)