Overview¶
Equilibrium Climate Sensitivity (ECS) is defined as change in global-mean near-surface air temperature (GMST) change due to an instantaneous doubling of CO concentrations and once the coupled ocean-atmosphere-sea ice system has acheived a statistical equilibrium (i.e. at the top-of-atmosphere, incoming solar shortwave radiation is balanced by reflected solar shortwave and outgoing thermal longwave radiation).
This notebook uses the “Gregory method” to approximate the ECS of CMIP6 models based on the first 150 years after an abrupt doubling of CO concentrations. The Gregory method extrapolates the quasi-linear relationship between GMST and radiative imbalance at the top-of-atmosphere to estimate how much warming would occur if the system were in radiative balance at the top-of-atmosphere, which is by definition the equilibrium response. In particular, we extrapolate the linear relationship that occurs between 100 and 150 years after the abrupt quadrupling.
Since the radiative forcing due to CO is a logarithmic function of the CO concentration, the GMST change from a first doubling is roughly the same as for a second doubling (to first order, we can assume feedbacks as constant), which means that the GMST change due to a quadrupling of CO is roughly . See also Mauritsen et al. 2019 for a detailed application of the Gregory method (with modifications) for the case of one specific CMIP6 model, the MPI-M Earth System Model.
For another take on applying the Gregory method to estimate ECS, see Angeline Pendergrass’ code.
Prerequisites¶
| Concepts | Importance | Notes |
|---|---|---|
| Intro to Xarray | Necessary | |
| Understanding of NetCDF | Helpful | Familiarity with metadata structure |
| Dask | Helpful | |
| Climate sensitivity | Helpful |
- Time to learn: 30 minutes
Imports¶
from matplotlib import pyplot as plt
import sys
import numpy as np
import pandas as pd
import xarray as xr
import cartopy
import dask
from tqdm.autonotebook import tqdm # Fancy progress bars for our loops!
import intake
import fsspec
from dask_gateway import Gateway
from dask.distributed import Client
%matplotlib inline
plt.rcParams['figure.figsize'] = 12, 6/tmp/ipykernel_4302/1804016931.py:8: TqdmExperimentalWarning: Using `tqdm.autonotebook.tqdm` in notebook mode. Use `tqdm.tqdm` instead to force console mode (e.g. in jupyter console)
from tqdm.autonotebook import tqdm # Fancy progress bars for our loops!
Compute Cluster¶
Here we use a dask cluster to parallelize our analysis.
platform = sys.platform
if (platform == 'win32'):
import multiprocessing.popen_spawn_win32
else:
import multiprocessing.popen_spawn_posixInitiate the Dask client:
client = Client()
clientData catalogs¶
This notebook uses intake-esm to ingest and organize climate model output from the fresh-off-the-supercomputers Phase 6 of the Coupled Model Intercomparison Project (CMIP6).
The file https://storage.googleapis.com/cmip6/cmip6-zarr-consolidated-stores.csv in Google Cloud Storage contains thousands of lines of metadata, each describing an individual climate model experiment’s simulated data.
For example, the first line in the .csv file contains the precipitation rate (variable_id = 'pr'), as a function of latitude, longitude, and time, in an individual climate model experiment with the BCC-ESM1 model (source_id = 'BCC-ESM1') developed by the Beijing Climate Center (institution_id = 'BCC'). The model is forced by the forcing experiment SSP370 (experiment_id = 'ssp370'), which stands for the Shared Socio-Economic Pathway 3 that results in a change in radiative forcing of W m from pre-industrial to 2100. This simulation was run as part of the AerChemMIP activity, which is a spin-off of the CMIP activity that focuses specifically on how aerosol chemistry affects climate.
df = pd.read_csv('https://storage.googleapis.com/cmip6/cmip6-zarr-consolidated-stores.csv')
df.head()The file pangeo-cmip6.json describes the structure of the CMIP6 metadata and is formatted so as to be read in by the intake.open_esm_datastore method, which categorizes all of the data pointers into a tiered collection. For example, this collection contains the simulated data from 28691 individual experiments, representing 48 different models from 23 different scientific institutions. There are 190 different climate variables (e.g. sea surface temperature, sea ice concentration, atmospheric winds, dissolved organic carbon in the ocean, etc.) available for 29 different forcing experiments.
Use Intake-ESM¶
Intake-ESM is a new package designed to make working with these data archives a bit simpler.
col = intake.open_esm_datastore("https://storage.googleapis.com/cmip6/pangeo-cmip6.json")
colHere, we show the various forcing experiments that climate modellers ran in these simulations.
df['experiment_id'].unique()array(['highresSST-present', 'piControl', 'control-1950', 'hist-1950',
'historical', 'amip', 'abrupt-4xCO2', 'abrupt-2xCO2',
'abrupt-0p5xCO2', '1pctCO2', 'ssp585', 'esm-piControl', 'esm-hist',
'hist-piAer', 'histSST-1950HC', 'ssp245', 'hist-1950HC', 'histSST',
'piClim-2xVOC', 'piClim-2xNOx', 'piClim-2xdust', 'piClim-2xss',
'piClim-histall', 'hist-piNTCF', 'histSST-piNTCF',
'aqua-control-lwoff', 'piClim-lu', 'histSST-piO3', 'piClim-CH4',
'piClim-NTCF', 'piClim-NOx', 'piClim-O3', 'piClim-HC',
'faf-heat-NA0pct', 'ssp370SST-lowCH4', 'piClim-VOC',
'ssp370-lowNTCF', 'piClim-control', 'piClim-aer', 'hist-aer',
'faf-heat', 'faf-heat-NA50pct', 'ssp370SST-lowNTCF',
'ssp370SST-ssp126Lu', 'ssp370SST', 'ssp370pdSST', 'histSST-piAer',
'piClim-ghg', 'piClim-anthro', 'faf-all', 'hist-nat', 'hist-GHG',
'ssp119', 'piClim-histnat', 'piClim-4xCO2', 'ssp370',
'piClim-histghg', 'highresSST-future', 'esm-ssp585-ssp126Lu',
'ssp126-ssp370Lu', 'ssp370-ssp126Lu', 'land-noLu', 'histSST-piCH4',
'ssp126', 'esm-pi-CO2pulse', 'amip-hist', 'piClim-histaer',
'amip-4xCO2', 'faf-water', 'faf-passiveheat', '1pctCO2-rad',
'faf-stress', '1pctCO2-bgc', 'aqua-control', 'amip-future4K',
'amip-p4K', 'aqua-p4K', 'amip-lwoff', 'amip-m4K', 'aqua-4xCO2',
'amip-p4K-lwoff', 'hist-noLu', '1pctCO2-cdr',
'land-hist-altStartYear', 'land-hist', 'omip1', 'esm-pi-cdr-pulse',
'esm-ssp585', 'abrupt-solp4p', 'piControl-spinup', 'hist-stratO3',
'abrupt-solm4p', 'midHolocene', 'lig127k', 'aqua-p4K-lwoff',
'esm-piControl-spinup', 'ssp245-GHG', 'ssp245-nat',
'dcppC-amv-neg', 'dcppC-amv-ExTrop-neg', 'dcppC-atl-control',
'dcppC-amv-pos', 'dcppC-ipv-NexTrop-neg', 'dcppC-ipv-NexTrop-pos',
'dcppC-atl-pacemaker', 'dcppC-amv-ExTrop-pos',
'dcppC-amv-Trop-neg', 'dcppC-pac-control', 'dcppC-ipv-pos',
'dcppC-pac-pacemaker', 'dcppC-ipv-neg', 'dcppC-amv-Trop-pos',
'piClim-BC', 'piClim-2xfire', 'piClim-SO2', 'piClim-OC',
'piClim-N2O', 'piClim-2xDMS', 'ssp460', 'ssp434', 'ssp534-over',
'deforest-globe', 'historical-cmip5', 'hist-bgc',
'piControl-cmip5', 'rcp26-cmip5', 'rcp45-cmip5', 'rcp85-cmip5',
'pdSST-piArcSIC', 'pdSST-piAntSIC', 'piSST-piSIC', 'piSST-pdSIC',
'ssp245-stratO3', 'hist-sol', 'hist-CO2', 'hist-volc',
'hist-totalO3', 'hist-nat-cmip5', 'hist-aer-cmip5',
'hist-GHG-cmip5', 'pdSST-futAntSIC', 'futSST-pdSIC', 'pdSST-pdSIC',
'ssp245-aer', 'pdSST-futArcSIC', 'dcppA-hindcast', 'dcppA-assim',
'dcppC-hindcast-noPinatubo', 'dcppC-hindcast-noElChichon',
'dcppC-hindcast-noAgung', 'hist-resIPO', 'ssp245-cov-modgreen',
'ssp245-cov-fossil', 'ssp245-cov-strgreen', 'ssp245-covid', 'lgm',
'ssp585-bgc', '1pctCO2to4x-withism', '1pctCO2-4xext', 'past1000',
'pa-futArcSIC', 'pa-pdSIC', 'historical-ext', 'pdSST-futArcSICSIT',
'pdSST-futOkhotskSIC', 'pdSST-futBKSeasSIC', 'pa-piArcSIC',
'pa-piAntSIC', 'pa-futAntSIC', 'pdSST-pdSICSIT'], dtype=object)Loading Data¶
Intake-ESM enables loading data directly into an xarray.DataArray, a metadata-aware extension of numpy arrays. Xarray objects leverage Dask to only read data into memory as needed for any specific operation (i.e. lazy evaluation). Think of Xarray Datasets as ways of conveniently organizing large arrays of floating point numbers (e.g. climate model data) on an n-dimensional discrete grid, with important metadata such as units, variable, names, etc.
Note that data on the cloud are in Zarr format, an extension of the metadata-aware format NetCDF commonly used in the geosciences.
Intake-ESM has rules for aggegating datasets; these rules are defined in the collection-specification file.
Here, we choose the piControl experiment (in which CO concentrations are held fixed at a pre-industrial level of ~300 ppm) and abrupt-2xCO2 experiment (in which CO concentrations are instantaneously doubled from a pre-industrial control state). Since the radiative forcing of CO is roughly a logarithmic function of CO concentrations, the ECS is roughly independent of the initial CO concentration.
Warning
The version of this notebook in the Pangeo Gallery uses theabrupt-4xCO2 forcing experiment, but fewer abrupt-2xCO2 datasets are currently avaiable in Google Cloud Storage, which significantly reduces run time. If you want to run this notebook on your own computer with the abrupt-4xCO2 experiment instead, change co2_option in the cell below. You will also need to take half of ecs, as described in the Overview.co2_option = 'abrupt-2xCO2'Prepare Data¶
query = dict(
experiment_id=[co2_option,'piControl'], # pick the `abrupt-2xCO2` and `piControl` forcing experiments
table_id='Amon', # choose to look at atmospheric variables (A) saved at monthly resolution (mon)
variable_id=['tas', 'rsut','rsdt','rlut'], # choose to look at near-surface air temperature (tas) as our variable
member_id = 'r1i1p1f1', # arbitrarily pick one realization for each model (i.e. just one set of initial conditions)
)
col_subset = col.search(require_all_on=["source_id"], **query)
col_subset.df.groupby("source_id")[
["experiment_id", "variable_id", "table_id"]
].nunique()The following functions help us load and homogenize the data. We use some dask.delayed programming to open the datasets in parallel.
def drop_all_bounds(ds):
"""Drop coordinates like 'time_bounds' from datasets,
which can lead to issues when merging."""
drop_vars = [vname for vname in ds.coords
if (('_bounds') in vname ) or ('_bnds') in vname]
return ds.drop_vars(drop_vars)
def open_dsets(df):
"""Open datasets from cloud storage and return xarray dataset."""
dsets = [xr.open_zarr(fsspec.get_mapper(ds_url), consolidated=True)
.pipe(drop_all_bounds)
for ds_url in df.zstore]
try:
ds = xr.merge(dsets, join='exact')
return ds
except ValueError:
return None
def open_delayed(df):
"""A dask.delayed wrapper around `open_dsets`.
Allows us to open many datasets in parallel."""
return dask.delayed(open_dsets)(df)Create a nested dictionary of models and experiments. It will be structured like this:
{'CESM2':
{
'piControl': <xarray.Dataset>,
'abrupt-2xCO2': <xarray.Dataset>
},
...
}from collections import defaultdict
dsets = defaultdict(dict)
for group, df in col_subset.df.groupby(by=['source_id', 'experiment_id']):
dsets[group[0]][group[1]] = open_delayed(df)Open one of the datasets directly, just to show what it looks like:
%time open_dsets(df)Now use dask to do this in parallel on all of the datasets:
dsets_ = dask.compute(dict(dsets))[0]/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
/tmp/ipykernel_4302/1448205721.py:10: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
Reduce Data via Global Mean¶
We don’t want to load all of the raw model data into memory right away. Instead, we want to reduce the data by taking the global mean. We need to remember to weight this global mean by a factor proportional to cos(lat).
def get_lat_name(ds):
"""Figure out what is the latitude coordinate for each dataset."""
for lat_name in ['lat', 'latitude']:
if lat_name in ds.coords:
return lat_name
raise RuntimeError("Couldn't find a latitude coordinate")
def global_mean(ds):
"""Return global mean of a whole dataset."""
lat = ds[get_lat_name(ds)]
weight = np.cos(np.deg2rad(lat))
weight /= weight.mean()
other_dims = set(ds.dims) - {'time'}
return (ds * weight).mean(other_dims)We now apply this function, plus resampling to annual mean data, to all of the datasets. We also concatenate the experiments together into a single Dataset for each model. This is the most complex cell in the notebook. A lot is happening here.
expts = ['piControl', co2_option]
expt_da = xr.DataArray(expts, dims='experiment_id',
coords={'experiment_id': expts})
dsets_aligned = {}
for k, v in tqdm(dsets_.items()):
expt_dsets = v.values()
if any([d is None for d in expt_dsets]):
print(f"Missing experiment for {k}")
continue
for ds in expt_dsets:
ds.coords['year'] = ds.time.dt.year - ds.time.dt.year[0]
# workaround for
# https://github.com/pydata/xarray/issues/2237#issuecomment-620961663
dsets_ann_mean = [v[expt].pipe(global_mean)
.swap_dims({'time': 'year'})
.drop_vars('time')
.coarsen(year=12).mean()
for expt in expts]
# align everything with the 2xCO2 experiment
dsets_aligned[k] = xr.concat(dsets_ann_mean, join='right',
dim=expt_da)Do the Computation¶
Up to this point, no computations have actually happened. Everything has been “lazy”. Now we trigger the computation to actual occur and load the global/annual mean data into memory.
dsets_aligned_ = dask.compute(dsets_aligned)[0]Now we concatenate across models to produce one big dataset with all the required variables.
source_ids = list(dsets_aligned_.keys())
source_da = xr.DataArray(source_ids, dims='source_id',
coords={'source_id': source_ids})
big_ds = xr.concat([ds.reset_coords(drop=True)
for ds in dsets_aligned_.values()],
dim=source_da)
big_dsCalculated Derived Variables¶
We need to calculate the net radiative imbalance, plus the anomaly of the abrupt 2xCO2 run compared to the piControl run.
big_ds['imbalance'] = big_ds['rsdt'] - big_ds['rsut'] - big_ds['rlut']
ds_mean = big_ds[['tas', 'imbalance']].sel(experiment_id='piControl').mean(dim='year')
ds_anom = big_ds[['tas', 'imbalance']] - ds_mean
# add some metadata
ds_anom.tas.attrs['long_name'] = 'Global Mean Surface Temp Anom'
ds_anom.tas.attrs['units'] = 'K'
ds_anom.imbalance.attrs['long_name'] = 'Global Mean Radiative Imbalance'
ds_anom.imbalance.attrs['units'] = 'W m$^{-2}$'
ds_anomPlot Timeseries¶
Here we plot the global mean surface temperature for each model:
ds_anom.tas.plot.line(col='source_id', x='year', col_wrap=5)<xarray.plot.facetgrid.FacetGrid at 0x7f814257b790>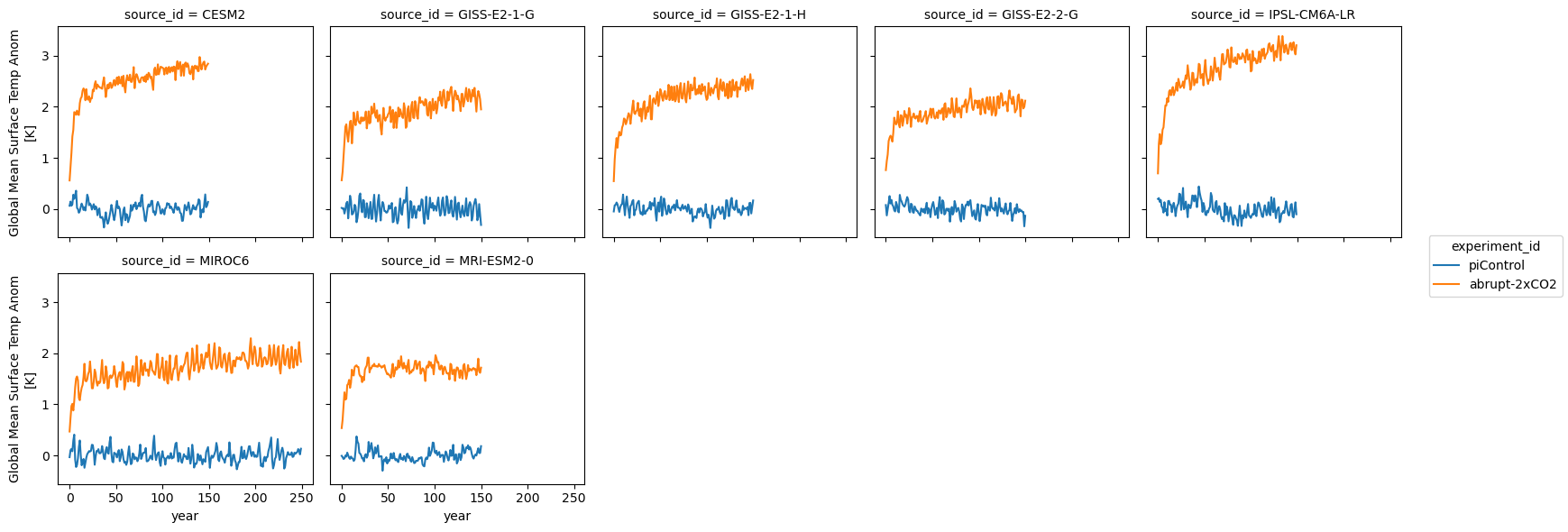
We can see that the models cover different time intervals. Let’s limit the rest of our analysis to the first 150 years.
first_150_years = slice(0, 149)
ds_anom.tas.sel(year=first_150_years).plot.line(col='source_id', x='year', col_wrap=5)<xarray.plot.facetgrid.FacetGrid at 0x7f81301ea6d0>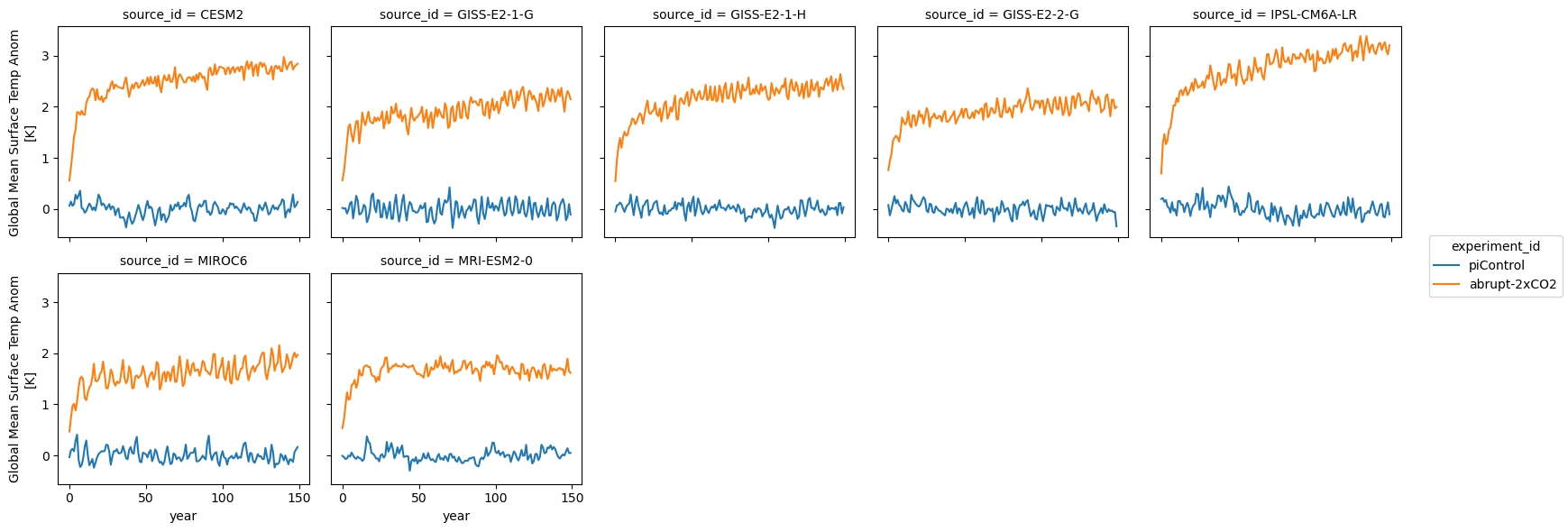
Same thing for radiative imbalance:
ds_anom.imbalance.sel(year=first_150_years).plot.line(col='source_id', x='year', col_wrap=5)<xarray.plot.facetgrid.FacetGrid at 0x7f81302c3790>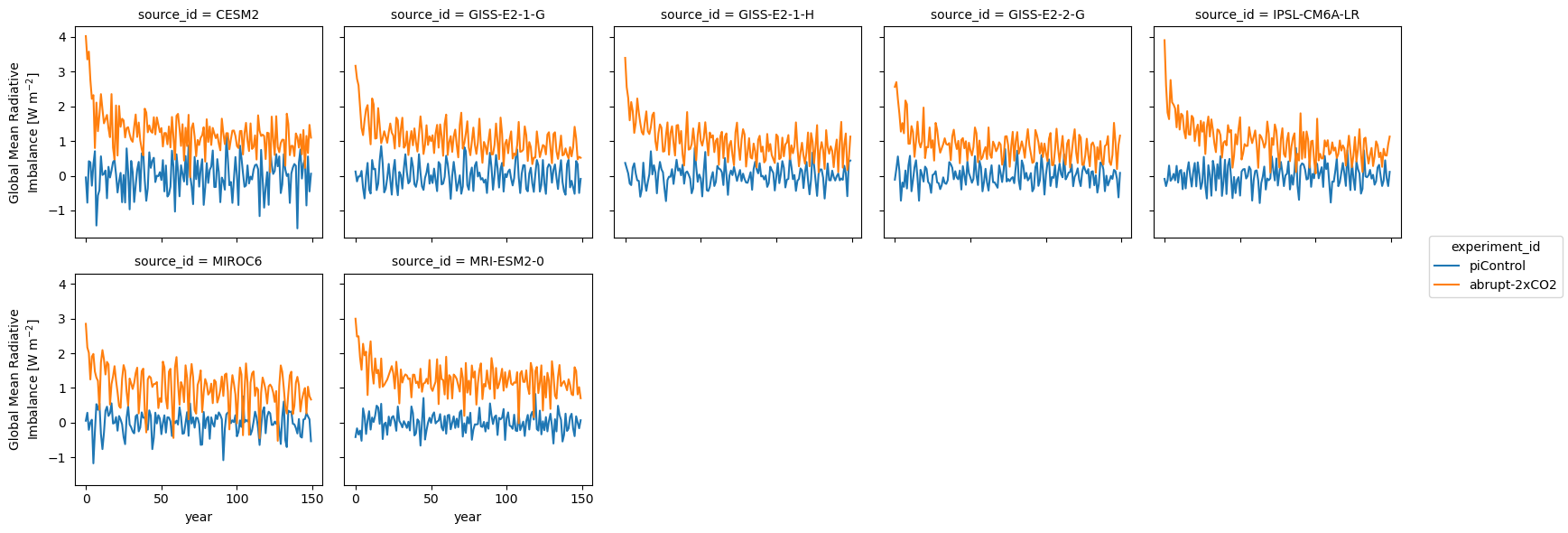
Calculate ECS¶
ds_abrupt = ds_anom.sel(year=first_150_years, experiment_id=co2_option).reset_coords(drop=True)
def calc_ecs(tas, imb):
a, b = np.polyfit(tas, imb, 1)
ecs = -1.0 * (b/a) # Change -1.0 to -0.5 if using 4xCO2
return xr.DataArray(ecs)
ds_abrupt['ecs'] = xr.apply_ufunc(calc_ecs, ds_abrupt.tas, ds_abrupt.imbalance, vectorize=True, input_core_dims=[['year'], ['year']])
ds_abrupt.compute()Also, make sure that we set a couple of the variables to be coordinates.
ds_abrupt = ds_abrupt.set_coords(['tas', 'imbalance'])
fg = ds_abrupt.plot.scatter(x='tas', y='imbalance', col='source_id', col_wrap=4, add_colorbar=False)
def calc_and_plot_ecs(x, y, **kwargs):
x = x[~np.isnan(x)]
y = y[~np.isnan(y)]
a, b = np.polyfit(x, y, 1)
ecs = -1.0 * b/a
plt.autoscale(False)
plt.plot([0, 10], np.polyval([a, b], [0, 10]), 'k')
plt.text(2, 3, f'ECS = {ecs:3.2f}', fontdict={'weight': 'bold', 'size': 12})
plt.grid()
fg.map(calc_and_plot_ecs, 'tas', 'imbalance')<xarray.plot.facetgrid.FacetGrid at 0x7f8124c821d0>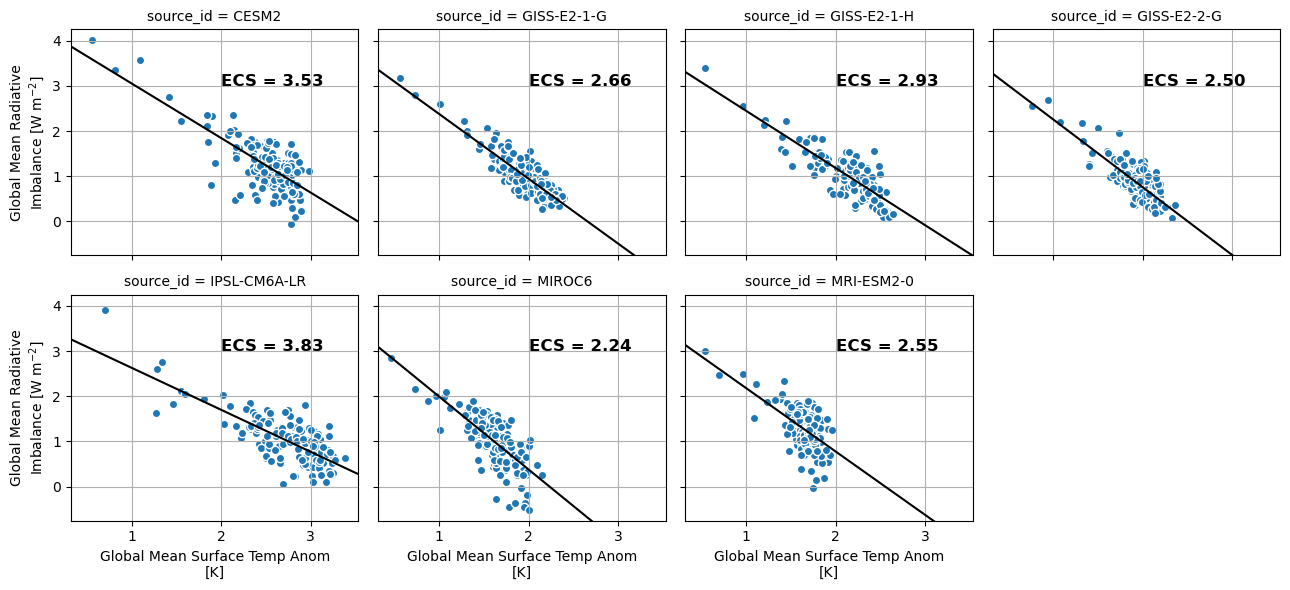
ds_abrupt.ecs.plot.hist();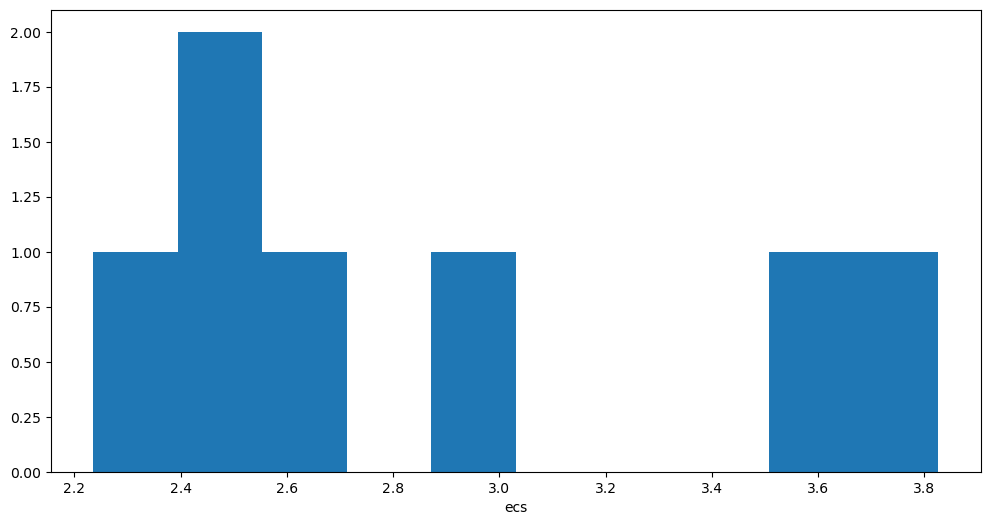
ds_abrupt.ecs.to_dataframe().sort_values('ecs').plot(kind='bar')<Axes: xlabel='source_id'>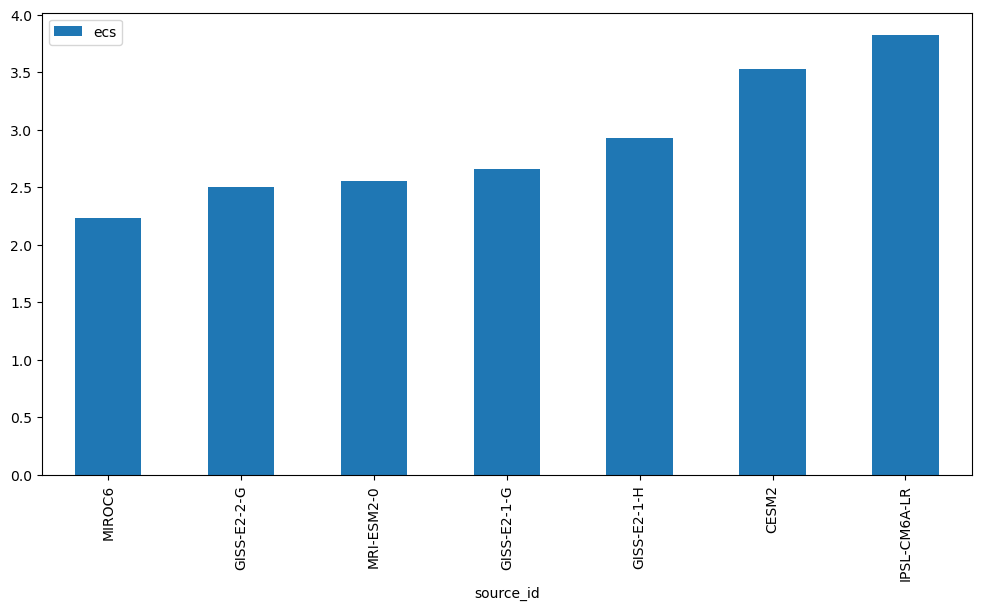
We’re at the end of the notebook, so let’s shutdown our Dask cluster.
client.shutdown()Summary¶
In this notebook, we estimated ECS for a subset of CMIP6 models using the Gregory method.
What’s next?¶
We will plot global average surface air temperature for a historical run and two branching emissions scenarios.
Resources and references¶
- Original notebook in the Pangeo Gallery by Henri Drake and Ryan Abernathey
- Gregory, J. M., Ingram, W. J., Palmer, M. A., Jones, G. S., Stott, P. A., Thorpe, R. B., Lowe, J. A., Johns, T. C., & Williams, K. D. (2004). A new method for diagnosing radiative forcing and climate sensitivity. Geophysical Research Letters, 31(3). 10.1029/2003gl018747
- Mauritsen, T., Bader, J., Becker, T., Behrens, J., Bittner, M., Brokopf, R., Brovkin, V., Claussen, M., Crueger, T., Esch, M., Fast, I., Fiedler, S., Fläschner, D., Gayler, V., Giorgetta, M., Goll, D. S., Haak, H., Hagemann, S., Hedemann, C., … Roeckner, E. (2019). Developments in the MPI‐M Earth System Model version 1.2 (MPI‐ESM1.2) and Its Response to Increasing CO2. Journal of Advances in Modeling Earth Systems, 11(4), 998–1038. 10.1029/2018ms001400