Overview¶
This notebook uses similar techniques to the ECS notebook. Please refer to that notebook for details.
Prerequisites¶
| Concepts | Importance | Notes |
|---|---|---|
| Understanding of NetCDF | Helpful | Familiarity with metadata structure |
| Seaborn | Helpful |
- Time to learn: 10 minutes
Imports¶
from matplotlib import pyplot as plt
import xarray as xr
import numpy as np
import dask
from dask.diagnostics import progress
from tqdm.autonotebook import tqdm
import intake
import fsspec
import seaborn as sns
%matplotlib inline/tmp/ipykernel_4226/1335193511.py:6: TqdmExperimentalWarning: Using `tqdm.autonotebook.tqdm` in notebook mode. Use `tqdm.tqdm` instead to force console mode (e.g. in jupyter console)
from tqdm.autonotebook import tqdm
col = intake.open_esm_datastore("https://storage.googleapis.com/cmip6/pangeo-cmip6.json")
colLoading...
[eid for eid in col.df['experiment_id'].unique() if 'ssp' in eid]['ssp585',
'ssp245',
'ssp370SST-lowCH4',
'ssp370-lowNTCF',
'ssp370SST-lowNTCF',
'ssp370SST-ssp126Lu',
'ssp370SST',
'ssp370pdSST',
'ssp119',
'ssp370',
'esm-ssp585-ssp126Lu',
'ssp126-ssp370Lu',
'ssp370-ssp126Lu',
'ssp126',
'esm-ssp585',
'ssp245-GHG',
'ssp245-nat',
'ssp460',
'ssp434',
'ssp534-over',
'ssp245-stratO3',
'ssp245-aer',
'ssp245-cov-modgreen',
'ssp245-cov-fossil',
'ssp245-cov-strgreen',
'ssp245-covid',
'ssp585-bgc']There is currently a significant amount of data for these runs:
expts = ['historical', 'ssp245', 'ssp585']
query = dict(
experiment_id=expts,
table_id='Amon',
variable_id=['tas'],
member_id = 'r1i1p1f1',
)
col_subset = col.search(require_all_on=["source_id"], **query)
col_subset.df.groupby("source_id")[
["experiment_id", "variable_id", "table_id"]
].nunique()Loading...
def drop_all_bounds(ds):
drop_vars = [vname for vname in ds.coords
if (('_bounds') in vname ) or ('_bnds') in vname]
return ds.drop(drop_vars)
def open_dset(df):
assert len(df) == 1
ds = xr.open_zarr(fsspec.get_mapper(df.zstore.values[0]), consolidated=True)
return drop_all_bounds(ds)
def open_delayed(df):
return dask.delayed(open_dset)(df)
from collections import defaultdict
dsets = defaultdict(dict)
for group, df in col_subset.df.groupby(by=['source_id', 'experiment_id']):
dsets[group[0]][group[1]] = open_delayed(df)dsets_ = dask.compute(dict(dsets))[0]/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:8: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
ds = xr.open_zarr(fsspec.get_mapper(df.zstore.values[0]), consolidated=True)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:8: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
ds = xr.open_zarr(fsspec.get_mapper(df.zstore.values[0]), consolidated=True)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:8: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
ds = xr.open_zarr(fsspec.get_mapper(df.zstore.values[0]), consolidated=True)
/tmp/ipykernel_4226/354912406.py:8: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
ds = xr.open_zarr(fsspec.get_mapper(df.zstore.values[0]), consolidated=True)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:8: SerializationWarning: Unable to decode time axis into full numpy.datetime64[ns] objects, continuing using cftime.datetime objects instead, reason: dates out of range. To silence this warning use a coarser resolution 'time_unit' or specify 'use_cftime=True'.
ds = xr.open_zarr(fsspec.get_mapper(df.zstore.values[0]), consolidated=True)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
/tmp/ipykernel_4226/354912406.py:4: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
return ds.drop(drop_vars)
Calculate global means:
def get_lat_name(ds):
for lat_name in ['lat', 'latitude']:
if lat_name in ds.coords:
return lat_name
raise RuntimeError("Couldn't find a latitude coordinate")
def global_mean(ds):
lat = ds[get_lat_name(ds)]
weight = np.cos(np.deg2rad(lat))
weight /= weight.mean()
other_dims = set(ds.dims) - {'time'}
return (ds * weight).mean(other_dims)expt_da = xr.DataArray(expts, dims='experiment_id', name='experiment_id',
coords={'experiment_id': expts})
dsets_aligned = {}
for k, v in tqdm(dsets_.items()):
expt_dsets = v.values()
if any([d is None for d in expt_dsets]):
print(f"Missing experiment for {k}")
continue
for ds in expt_dsets:
ds.coords['year'] = ds.time.dt.year
# workaround for
# https://github.com/pydata/xarray/issues/2237#issuecomment-620961663
dsets_ann_mean = [v[expt].pipe(global_mean)
.swap_dims({'time': 'year'})
.drop('time')
.coarsen(year=12).mean()
for expt in expts]
# align everything with the 4xCO2 experiment
dsets_aligned[k] = xr.concat(dsets_ann_mean, join='outer',
dim=expt_da)Loading...
dsets_aligned_ = dask.compute(dsets_aligned)[0]source_ids = list(dsets_aligned_.keys())
source_da = xr.DataArray(source_ids, dims='source_id', name='source_id',
coords={'source_id': source_ids})
big_ds = xr.concat([ds.reset_coords(drop=True)
for ds in dsets_aligned_.values()],
dim=source_da)
big_dsLoading...
df_all = big_ds.sel(year=slice(1900, 2100)).to_dataframe().reset_index()
df_all.head()Loading...
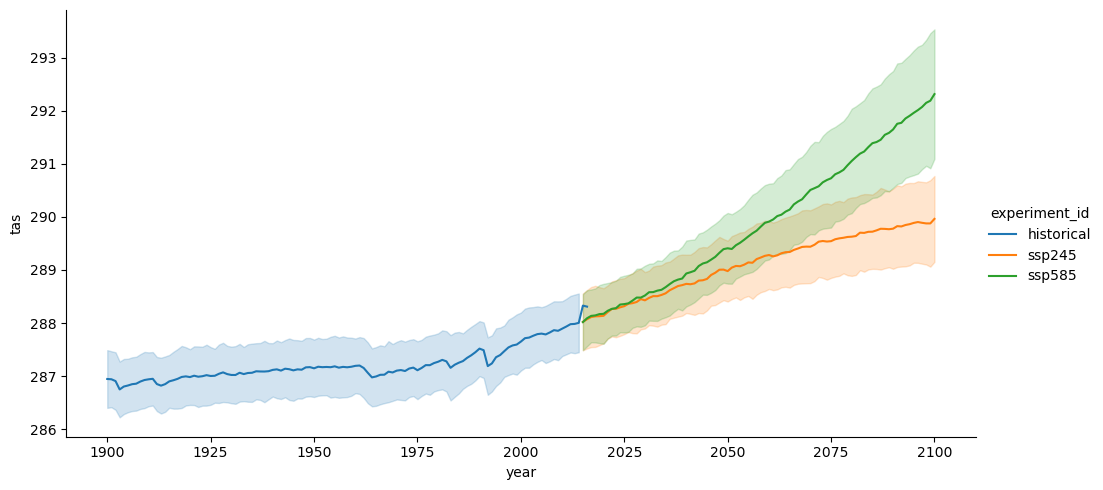
sns.relplot(data=df_all,
x="year", y="tas", hue='experiment_id',
kind="line", ci="sd", aspect=2);/home/runner/micromamba/envs/cmip6-cookbook-dev/lib/python3.11/site-packages/seaborn/axisgrid.py:854: FutureWarning:
The `ci` parameter is deprecated. Use `errorbar='sd'` for the same effect.
func(*plot_args, **plot_kwargs)
Summary¶
In this notebook, we accessed data for historical, SSP245, and SSP585 runs from a collection of CMIP6 models and plotted the multimodel-mean global average surface air temperature for each run.
What’s next?¶
We will use CMIP6 data to analyze precipitation intensity under a warming climate.
Resources and references¶
- Original notebook in the Pangeo Gallery by Henri Drake and Ryan Abernathey