
easy.gems for HEALPix Analysis & Visualization¶
In this section, you’ll learn:¶
Utilizing intake to open a HEALPix data catalog
Using the
healpixpackage to perform HEALPix operations to look at basic statisticsPlotting HEALPix data via easy.gems functionality
Related Documentation¶
Prerequisites¶
| Concepts | Importance | Notes |
|---|---|---|
| HEALPix overview | Necessary |
Time to learn: 30 minutes
Open data catalog¶
Let us use the online data catalog from the WCRP’s Digital Earths Global Hackathon 2025’s catalog repository using intake and read the output of the ICON simulation run ngc4008, which is stored in the HEALPix format:
import intake
# Hackathon data catalogs
cat_url = "https://digital-earths-global-hackathon.github.io/catalog/catalog.yaml"
cat = intake.open_catalog(cat_url).online
model_run = cat.icon_ngc4008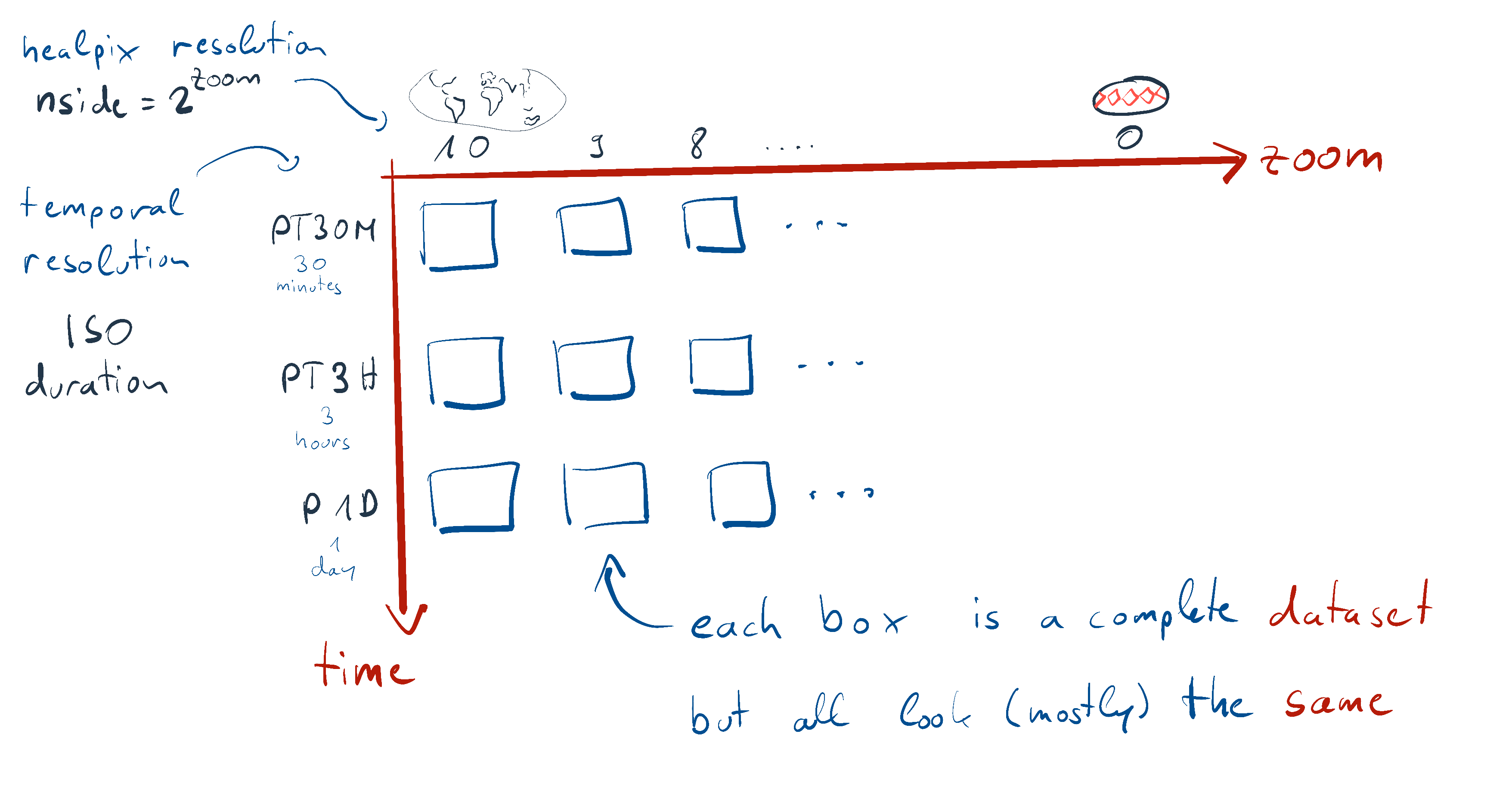
Explore datasets¶
So, the coarsest dataset in this model run would be as follows (Even if we called it without specifying any parameters, i.e. model_run.to_dask(), the result would be same as the ds_coarsest below since this model run defaults to the coarsest settings):
ds_coarsest = model_run(zoom=0, time="P1D").to_dask()
ds_coarsest/home/runner/micromamba/envs/healpix-cookbook-dev/lib/python3.13/site-packages/intake_xarray/base.py:21: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
'dims': dict(self._ds.dims),
Now, let us look at a dataset with finer zoom level still with the coarsest time and another dataset with a finer zoom level and the finest time (which may be useful for daily analyses) dataset:
ds_fine = model_run(zoom=7).to_dask()
ds_fine/home/runner/micromamba/envs/healpix-cookbook-dev/lib/python3.13/site-packages/intake_xarray/base.py:21: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
'dims': dict(self._ds.dims),
ds_finesttime = model_run(zoom=6, time="PT15M").to_dask()
ds_finesttime/home/runner/micromamba/envs/healpix-cookbook-dev/lib/python3.13/site-packages/intake_xarray/base.py:21: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
'dims': dict(self._ds.dims),
HEALPix basic stats with thehealpix package¶
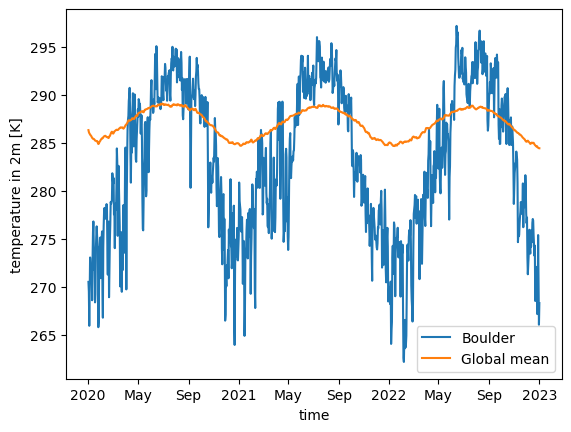
Let us look at the global and Boulder, CO, USA temperature averages for a 3-year time-slice of the whole dataset.
For this, we will first need to define a few HEALPix helper functions to get the nest and nside values from the dataset, then find the HEALPix pixel that Boulder coords fall in, and finally plot those temporal averages using matplotlib.
import healpix as hp
import matplotlib.pylab as pltHEALPix helper functions¶
def get_nest(ds):
return ds.crs.healpix_order == "nest"
def get_nside(ds):
return ds.crs.healpix_nsideHEALPix pixel containing Boulder’s coords¶
%%time
boulder_lon = -105.2747
boulder_lat = 40.0190
boulder_pixel = hp.ang2pix(
get_nside(ds_fine), boulder_lon, boulder_lat, lonlat=True, nest=get_nest(ds_fine)
)CPU times: user 413 μs, sys: 0 ns, total: 413 μs
Wall time: 418 μs
Global and Boulder’s temperature averages¶
%%time
time_slice = slice("2020-01-02T00:00:00.000000000", "2023-01-01T00:00:00.000000000")
ds_fine.tas.sel(time=time_slice).isel(cell=boulder_pixel).plot(label="Boulder")
ds_coarsest.tas.sel(time=time_slice).mean("cell").plot(label="Global mean")
plt.legend()CPU times: user 360 ms, sys: 216 ms, total: 577 ms
Wall time: 1.82 s
Plotting with easy.gems and cartopy¶
In this part, we will look at the healpix_show function that is provided by easy.gems for convenient HEALPix plotting.
import easygems.healpix as eghp
import cartopy.crs as ccrs
import cartopy.feature as cf
import cmoceanGlobal plots¶
Most of this is matplotlib and cartopy code, but have a look at how eghp.healpix_show() is called. The following code will plot global temperature (at the first timestep for simplicity)
projection = ccrs.Robinson(central_longitude=-135.5808361)
fig, ax = plt.subplots(
figsize=(8, 4), subplot_kw={"projection": projection}, constrained_layout=True
)
ax.set_global()
eghp.healpix_show(ds_fine.tas.isel(time=0), ax=ax, cmap=cmocean.cm.thermal)
ax.add_feature(cf.COASTLINE, linewidth=0.8)
ax.add_feature(cf.BORDERS, linewidth=0.4)---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
Cell In[10], line 9
3 fig, ax = plt.subplots(
4 figsize=(8, 4), subplot_kw={"projection": projection}, constrained_layout=True
5 )
7 ax.set_global()
----> 9 eghp.healpix_show(ds_fine.tas.isel(time=0), ax=ax, cmap=cmocean.cm.thermal)
11 ax.add_feature(cf.COASTLINE, linewidth=0.8)
12 ax.add_feature(cf.BORDERS, linewidth=0.4)
File ~/micromamba/envs/healpix-cookbook-dev/lib/python3.13/site-packages/easygems/healpix/__init__.py:172, in healpix_show(var, method, nest, **kwargs)
171 def healpix_show(var, method="nearest", nest=True, **kwargs):
--> 172 r = HEALPixResampler(nside=get_nside(var), nest=nest, method=method)
174 return map_show(var, resampler=r, **kwargs)
File ~/micromamba/envs/healpix-cookbook-dev/lib/python3.13/site-packages/easygems/healpix/__init__.py:27, in get_nside(dx)
25 def get_nside(dx):
26 try:
---> 27 grid_mapping = dx.cf["grid_mapping"]
28 except AttributeError:
29 if dx.squeeze().ndim > 1:
File ~/micromamba/envs/healpix-cookbook-dev/lib/python3.13/site-packages/cf_xarray/accessor.py:3406, in CFDataArrayAccessor.__getitem__(self, key)
3401 if not isinstance(key, Hashable):
3402 raise KeyError(
3403 f"Cannot use an Iterable of keys with DataArrays. Expected a single string. Received {key!r} instead."
3404 )
-> 3406 return _getitem(self, key)
File ~/micromamba/envs/healpix-cookbook-dev/lib/python3.13/site-packages/cf_xarray/accessor.py:1241, in _getitem(accessor, key, skip)
1238 return ds.set_coords(coords)
1240 except KeyError:
-> 1241 raise KeyError(
1242 f"{kind}.cf does not understand the key {k!r}. "
1243 f"Use 'repr({kind}.cf)' (or '{kind}.cf' in a Jupyter environment) to see a list of key names that can be interpreted."
1244 ) from None
KeyError: "DataArray.cf does not understand the key 'grid_mapping'. Use 'repr(DataArray.cf)' (or 'DataArray.cf' in a Jupyter environment) to see a list of key names that can be interpreted."
Regional plots¶
If plotting a region of interest is desired, it is also possible through setting extents of the matplotlib plot.
Let us look into USA map using the Boulder, CO, USA coords we had used before for simplicity:
projection = ccrs.Robinson(central_longitude=boulder_lon)
fig, ax = plt.subplots(
figsize=(8, 4), subplot_kw={"projection": projection}, constrained_layout=True
)
ax.set_extent([boulder_lon-20, boulder_lon+40, boulder_lat-20, boulder_lat+10], crs=ccrs.PlateCarree())
eghp.healpix_show(ds_fine.tas.isel(time=0), ax=ax, cmap=cmocean.cm.thermal)
ax.add_feature(cf.COASTLINE, linewidth=0.8)
ax.add_feature(cf.BORDERS, linewidth=0.4)Further easy.gems and healpix¶
These are only a sampling of HEALPix and easy.gems capabilities; if you are interested in learning more, be sure to check out the usage examples at the easy.gems HEALPix Documentation.
What is next?¶
The next section will provide an UXarray workflow that loads in and analyzes & visualizes HEALPix data.