UXarray for Basic HEALPix Statistics & Visualization
In this section, you’ll learn:
Utilizing
intaketo open a HEALPix data catalogUsing the
uxarraypackage to look at basic statistics over HEALPix dataUsing UXarray plotting functionality on HEALPix data
Prerequisites
Concepts |
Importance |
Notes |
|---|---|---|
Necessary |
||
Necessary |
Time to learn: 30 minutes
import cartopy.crs as ccrs
import uxarray as ux
Open data catalog
Note
If you think that you first need to learn about Intake, Pythia’s Intake Cookbook is a great resource to do so.
Let us use the online data catalog from the WCRP’s Digital Earths Global Hackathon 2025’s catalog repository using intake and read the output of the ICON simulation run ngc4008, which is stored in the HEALPix format:
Note
This section uses the same data and will showcase similar operations as the previous section, e.g. basic statistics and global and regional data plotting, except at the end where further grid exploration methods will be demonstrated.
import intake
# Hackathon data catalogs
cat_url = "https://digital-earths-global-hackathon.github.io/catalog/catalog.yaml"
cat = intake.open_catalog(cat_url).online
model_run = cat.icon_ngc4008
We will be looking at two resolution levels, one is the coarsest zoom level of 0, which is the default in this model run, and the other is a finer one at the zoom level of 7:
ds_coarsest = model_run().to_dask()
ds_fine = model_run(zoom=7).to_dask()
/home/runner/miniconda3/envs/healpix-cookbook-dev/lib/python3.10/site-packages/intake_xarray/base.py:21: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
'dims': dict(self._ds.dims),
/home/runner/miniconda3/envs/healpix-cookbook-dev/lib/python3.10/site-packages/intake_xarray/base.py:21: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
'dims': dict(self._ds.dims),
Create UXarray Datasets from HEALPix
Now let us use those xarray.Datasets from the model run to open unstructured grid-aware uxarray.UxDataset:
%%time
uxds_coarsest = ux.UxDataset.from_healpix(ds_coarsest)
uxds_fine = ux.UxDataset.from_healpix(ds_fine)
uxds_fine
CPU times: user 21.4 ms, sys: 1.01 ms, total: 22.4 ms
Wall time: 22.2 ms
<xarray.UxDataset> Size: 14TB
Dimensions: (time: 10958, depth_half: 73,
n_face: 196608, level_full: 90,
crs: 1, depth_full: 72,
soil_depth_water_level: 5,
level_half: 91,
soil_depth_energy_level: 5)
Coordinates:
* crs (crs) float32 4B nan
* depth_full (depth_full) float32 288B 1.0 ... 5....
* depth_half (depth_half) float32 292B 0.0 ... 5....
* level_full (level_full) int32 360B 1 2 3 ... 89 90
* level_half (level_half) int32 364B 1 2 3 ... 90 91
* soil_depth_energy_level (soil_depth_energy_level) float32 20B ...
* soil_depth_water_level (soil_depth_water_level) float32 20B ...
* time (time) datetime64[ns] 88kB 2020-01-0...
Dimensions without coordinates: n_face
Data variables: (12/103)
A_tracer_v_to (time, depth_half, n_face) float32 629GB ...
FrshFlux_IceSalt (time, n_face) float32 9GB ...
FrshFlux_TotalIce (time, n_face) float32 9GB ...
Qbot (time, n_face) float32 9GB ...
Qtop (time, n_face) float32 9GB ...
Wind_Speed_10m (time, n_face) float32 9GB ...
... ...
vas (time, n_face) float32 9GB ...
w (time, depth_half, n_face) float32 629GB ...
wa_phy (time, level_half, n_face) float32 784GB ...
zg (level_full, n_face) float32 71MB ...
zghalf (level_half, n_face) float32 72MB ...
zos (time, n_face) float32 9GB ...HEALPix basic stats using UXarray
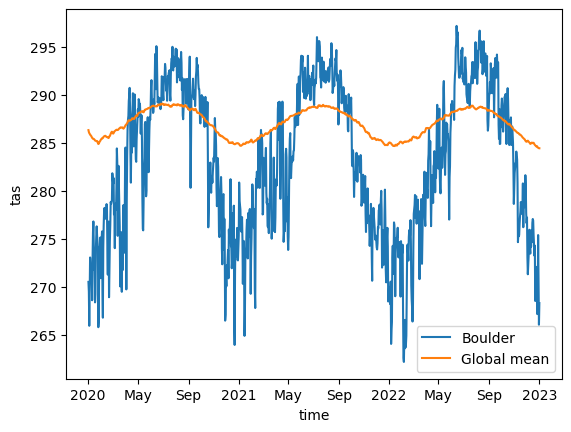
Let us look at the global and Boulder, CO, USA air temperature averages for the dataset. Data spans from 2020 to 2050, so let us also consider slicing it to have a 3-year interval between 2020 and 2023, which would also give us similar results to that with easy.gems in the previous section.
import matplotlib.pylab as plt
boulder_lon = -105.2747
boulder_lat = 40.0190
time_slice = slice("2020-01-02T00:00:00.000000000", "2023-01-01T00:00:00.000000000")
Mesh face containing Boulder’s coords
We can find face(s) containing a given point with uxarray conveniently as follows:
%%time
boulder_face = uxds_fine.uxgrid.get_faces_containing_point(
point_lonlat=[boulder_lon, boulder_lat]
)
CPU times: user 9 s, sys: 104 ms, total: 9.1 s
Wall time: 8.97 s
Data variables of interest
In order to use in the rest of the analyses, we can grab data variables, in theuxarray.UxDataArray type, from the dataset as follows:
uxda_fine = uxds_fine.tas
uxda_coarsest = uxds_coarsest.tas
Global and Boulder’s temperature averages
In order to get a line plot of our UXarray.UxDataset objects’ 1-dimensional temperature variables, we will convert them to xarray and call the default plot function because UXarray’s default plotting functions are all dedicated to grid-topology aware visualizations:
%%time
uxda_fine.isel(n_face=boulder_face).sel(time=time_slice).to_xarray().plot(
label="Boulder"
)
uxda_coarsest.sel(time=time_slice).mean("n_face").to_xarray().plot(label="Global mean")
plt.legend()
CPU times: user 353 ms, sys: 157 ms, total: 509 ms
Wall time: 2.34 s
<matplotlib.legend.Legend at 0x7f0362c75cc0>
Data plotting with UXarray
UXarray provides several built-in plotting functions to visualize unstructured grids, which can also be applied to HEALPix grids in the same interface:
Let us first look into interactive plots with the bokeh backend (i.e. UXarray’s plotting functions have a backend parameter that defaults to “bokeh”, and it can also accept “matplotlib”)
Global plots
Let us first plot the global temperature (at the first timestep for simplicity), using the default backend, bokeh, of UXarray’s visualization API to create an interactive plot:
%%time
projection = ccrs.Robinson(central_longitude=-135.5808361)
uxda_fine.isel(time=0).plot(
projection=projection,
cmap="inferno",
features=["borders", "coastline"],
title="Global temperature",
width=700,
)
CPU times: user 7.75 s, sys: 55 ms, total: 7.81 s
Wall time: 8.31 s
WARNING:param.GeoOverlayPlot00477: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
Now, let us create the same plot, using matplotlib as the backend:
%%time
uxda_fine.isel(time=0).plot(
backend="matplotlib",
projection=projection,
cmap="inferno",
features=["borders", "coastline"],
title="Global temperature",
width=1100,
)
CPU times: user 619 ms, sys: 28.9 ms, total: 648 ms
Wall time: 1.63 s
Regional subsets (Not only for plotting but also for analysis)
When a region on the globe is of interest, UXarray provides subsetting functions, which return new regional grids that can then be used in the same way a global grid is plotted.
Let us look into the USA map using the Boulder, CO, USA coords we had used before for simplicity:
Subsetting uxds_fine into a new UxDataset using a “bounding box” around Boulder, CO first:
%%time
lon_bounds = (boulder_lon - 20, boulder_lon + 40)
lat_bounds = (boulder_lat - 20, boulder_lat + 12)
uxda_fine_subset = uxda_fine.isel(time=0).subset.bounding_box(lon_bounds, lat_bounds)
/home/runner/miniconda3/envs/healpix-cookbook-dev/lib/python3.10/site-packages/uxarray/grid/grid.py:1432: RuntimeWarning: Necessary functions for computing the bounds of each face are not yet compiled with Numba. This initial execution will be significantly longer.
warn(
CPU times: user 1min 10s, sys: 154 ms, total: 1min 10s
Wall time: 1min 7s
If we check the global and regional subset’s average temperature at the first timestep, we can see the difference:
print(
"Global temperature average: ", uxda_fine.isel(time=0).mean("n_face").values, " K"
)
print(
"Regional subset's temperature average: ", uxda_fine_subset.mean("n_face").values, " K"
)
Global temperature average: 286.30957 K
Regional subset's temperature average: 281.8399 K
Now, let us plot the regional subset UxDataset:
%%time
projection = ccrs.Robinson(central_longitude=boulder_lon)
uxda_fine_subset.plot(
projection=projection,
cmap="inferno",
features=["borders", "coastline"],
title="Boulder temperature",
width=1100,
)
CPU times: user 73.2 ms, sys: 12 μs, total: 73.2 ms
Wall time: 72.7 ms
Grid topology exploration
Exploring the grid topology may be needed sometimes, and UXarray provides functionality to do so, both numerically and visually. Each UxDataset or UxDataArray has their associated Grid object that has all the information such as spherical and cartesian coordinates, connectives, dimensions, etc. about the topology this data belongs to. This Grid object can be explored as follows:
# uxds_fine.uxgrid # this would give the same as the below
uxda_fine.uxgrid
<uxarray.Grid> Original Grid Type: HEALPix Grid Dimensions: * n_node: 196610 * n_face: 196608 * n_max_face_nodes: 4 Grid Coordinates (Spherical): * node_lon: (196610,) * node_lat: (196610,) * face_lon: (196608,) * face_lat: (196608,) Grid Coordinates (Cartesian): * node_x: (196610,) * node_y: (196610,) * node_z: (196610,) Grid Connectivity Variables: * face_node_connectivity: (196608, 4) Grid Descriptor Variables: * n_nodes_per_face: (196608,)
There might be times that the user wants to open a standalone Grid object for a HEALPix grid (or any other unstructured grids supported by UXarray) without accessing the data yet. Let’s create the coarsest HEALPix grid as follows:
uxgrid = ux.Grid.from_healpix(zoom=0, pixels_only=False)
uxgrid
<uxarray.Grid> Original Grid Type: HEALPix Grid Dimensions: * n_node: 14 * n_face: 12 * n_max_face_nodes: 4 Grid Coordinates (Spherical): * node_lon: (14,) * node_lat: (14,) * face_lon: (12,) * face_lat: (12,) Grid Coordinates (Cartesian): Grid Connectivity Variables: * face_node_connectivity: (12, 4) Grid Descriptor Variables:
Let’s investigate how a HEALPix grid looks like over the poles. We can do things by selecting an Orthographic projection and setting a Geodetic source projection. This allows to better approximates the true HEALPix structured compared to the defaut PlateCaree projection.
projection = ccrs.Orthographic(central_latitude=90)
projection.threshold /= 100 # Smoothes out great circle arcs
uxgrid.plot(
periodic_elements="ignore", # Allow Cartopy to handle periodic elements
crs=ccrs.Geodetic(), # Enables edges to be plotted as GCAs
project=True,
projection=projection,
width=500,
title="HEALPix (Orthographic Proj), zoom=0",
)
Let’s chose another projection.
projection = ccrs.Mollweide()
projection.threshold /= 100 # Smoothes out great circle arcs
uxgrid.plot(
periodic_elements="ignore", # Allow Cartopy to handle periodic elements
crs=ccrs.Geodetic(), # Enables edges to be plotted as GCAs
project=True,
projection=projection,
width=500,
title="HEALPix (Mollweide Proj), zoom=0",
)
The grid structure here is approximated. While the boundary between each pixel is easily known in HEALPix workflows, UXarray represents the boundaries as a Great Circle Arc (GCA) due to the requirement of having explicit connectivity information, which is different than HEALPix boundaries and leads to minor differences in the computed plots and computations.
Now that we’ve looked at the grid structure, we can also apply the same principles to our data plotting.
Warning
Using the Geodetic source projection is not reccomended for higher-resolution grids, as it introduces a significant overhead.
uxda_coarsest.isel(time=0).plot(
periodic_elements="ignore", # Allow Cartopy to handle periodic elements
crs=ccrs.Geodetic(), # Enables edges to be plotted as GCAs
project=True,
projection=projection,
features=["borders", "coastline"],
cmap="inferno",
title="Temperature (Mollweide Proj), zoom=0",
width=500,
)
What is next?
The next section will provide an UXarray workflow that loads in and analyzes & visualizes HEALPix data.

