Basics of virtual Zarr stores
Overview
This notebook is intended as an introduction to creating and using virtual Zarr stores. In this tutorial we will:
Scan a single NetCDF4/HDF5 file to create a virtual dataset
Learn how to use the output using
XarrayandZarr.
While this notebook only examines using VirtualiZarr and Kerchunk on a single NetCDF file, these libraries can be used to create virtual Zarr datasets from collections of many input files. In the following notebook, we will demonstrate this.
Prerequisites
Concepts |
Importance |
Notes |
|---|---|---|
Helpful |
Basic features |
Time to learn: 60 minutes
Imports
Here we will import a few Python libraries to help with our data processing.
virtualizarrwill be used to generate the virtual Zarr storeXarrayfor examining the output dataset
import xarray as xr
from virtualizarr import open_virtual_dataset
Define storage_options arguments
In the dictionary definition in the next cell, we are defining the options that will be passed to fsspec.open. Any additional kwargs passed in this dictionary through fsspec.open will pass as kwargs to the file system, in our case s3. The API docs for the s3fs filesystem spec can be found here.
In this example we are passing a few kwargs. In short they are:
anon=True: This is as3fskwarg that specifies you are not passing any connection credentials and are connecting to a public bucket.default_fill_cache=False:s3fskwarg that avoids caching in between chunks of files. This may lower memory usage when reading large files.default_cache_type="none":fsspeckwarg that specifies the caching strategy used byfsspec. In this case, we turn off caching entirely to lower memory usage when only using the information from the file once…
storage_options = dict(anon=True, default_fill_cache=False, default_cache_type="none")
Virtualize a single NetCDF file
Below we will virtualize a NetCDF file stored on the AWS cloud. This dataset is a single time slice of a climate downscaled product for Alaska.
The steps in the cell below are as follows:
Create a virtual dataset using
open_virtual_datasetWrite the virtual store as a Kerchunk reference JSON using the
to_kerchunkmethod.
# Input URL to dataset. Note this is a netcdf file stored on s3 (cloud dataset).
url = "s3://wrf-se-ak-ar5/ccsm/rcp85/daily/2060/WRFDS_2060-01-01.nc"
# Create a virtual dataset using VirtualiZarr.
# We specify `indexes={}` to avoid creating in-memory pandas indexes for each 1D coordinate, since concatenating with pandas indexes is not yet supported in VirtualiZarr
virtual_ds = open_virtual_dataset(
url, indexes={}, reader_options={"storage_options": storage_options}
)
# Write the virtual dataset to disk as a Kerchunk JSON. We could alternative write to a Kerchunk JSON or Icechunk Store.
virtual_ds.virtualize.to_kerchunk("single_file_kerchunk.json", format="json")
Opening virtual datasets
In the section below we will use the previously created Kerchunk reference JSON to open the NetCDF file as if it were a Zarr dataset.
# We once again need to provide information for fsspec to access the remote file
storage_options = dict(
remote_protocol="s3", remote_options=dict(anon=True), skip_instance_cache=True
)
# We will use the "kerchunk" engine in `xr.open_dataset` and pass the `storage_options` to the `kerchunk` engine through `backend_kwargs`
ds = xr.open_dataset(
"single_file_kerchunk.json",
engine="kerchunk",
backend_kwargs={"storage_options": storage_options},
)
ds
<xarray.Dataset> Size: 31MB
Dimensions: (Time: 1, south_north: 250, west_east: 320,
interp_levels: 9, soil_layers_stag: 4)
Coordinates:
* Time (Time) datetime64[ns] 8B NaT
XLAT (south_north, west_east) float32 320kB ...
XLONG (south_north, west_east) float32 320kB ...
* interp_levels (interp_levels) float32 36B 100.0 200.0 300.0 ... 925.0 1e+03
Dimensions without coordinates: south_north, west_east, soil_layers_stag
Data variables: (12/37)
ACSNOW (Time, south_north, west_east) float32 320kB ...
ALBEDO (Time, south_north, west_east) float32 320kB ...
CLDFRA (Time, interp_levels, south_north, west_east) float32 3MB ...
GHT (Time, interp_levels, south_north, west_east) float32 3MB ...
HFX (Time, south_north, west_east) float32 320kB ...
LH (Time, south_north, west_east) float32 320kB ...
... ...
U (Time, interp_levels, south_north, west_east) float32 3MB ...
U10 (Time, south_north, west_east) float32 320kB ...
V (Time, interp_levels, south_north, west_east) float32 3MB ...
V10 (Time, south_north, west_east) float32 320kB ...
lat (south_north, west_east) float32 320kB ...
lon (south_north, west_east) float32 320kB ...
Attributes:
contact: rtladerjr@alaska.edu
data: Downscaled CCSM4
date: Mon Oct 21 11:37:23 AKDT 2019
format: version 2
info: Alaska CASCPlot dataset
ds.TMAX.plot()
<matplotlib.collections.QuadMesh at 0x7f6ffb532e60>
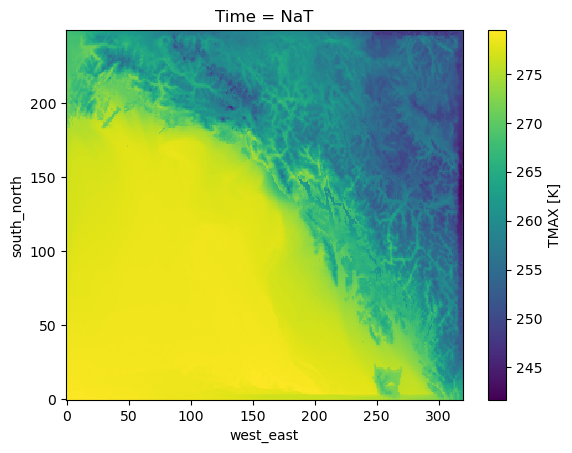
Note that the original .nc file size here is 16.8MB, and the created JSON is 26.5kB. These files also tend to compress very well. As you can see, it the JSON can be written anywhere, and gives us access to the underlying data, reading only the chunks we need from remote without downloading the whole file.