GeoTIFF
Generating virutal datasets from GeoTiff files
Overview
In this tutorial we will cover:
How to generate virtual datasets from GeoTIFFs.
Combining virtual datasets.
Prerequisites
Concepts |
Importance |
Notes |
|---|---|---|
Required |
Core |
|
Required |
Core |
|
Parallel virtual dataset creation with VirtualiZarr, Kerchunk, and Dask |
Required |
Core |
Required |
IO/Visualization |
Time to learn: 30 minutes
About the Dataset
The Finish Meterological Institute (FMI) Weather Radar Dataset is a collection of GeoTIFF files containing multiple radar specific variables, such as rainfall intensity, precipitation accumulation (in 1, 12 and 24 hour increments), radar reflectivity, radial velocity, rain classification and the cloud top height. It is available through the AWS public data portal and is updated frequently.
More details on this dataset can be found here.
import logging
from datetime import datetime
import dask
import fsspec
import rioxarray
import s3fs
import xarray as xr
from distributed import Client
from virtualizarr import open_virtual_dataset
Examining a Single GeoTIFF File
Before we use Kerchunk to create indices for multiple files, we can load a single GeoTiff file to examine it.
# URL pointing to a single GeoTIFF file
url = "s3://fmi-opendata-radar-geotiff/2023/07/01/FIN-ACRR-3067-1KM/202307010100_FIN-ACRR1H-3067-1KM.tif"
# Initialize a s3 filesystem
fs = s3fs.S3FileSystem(anon=True)
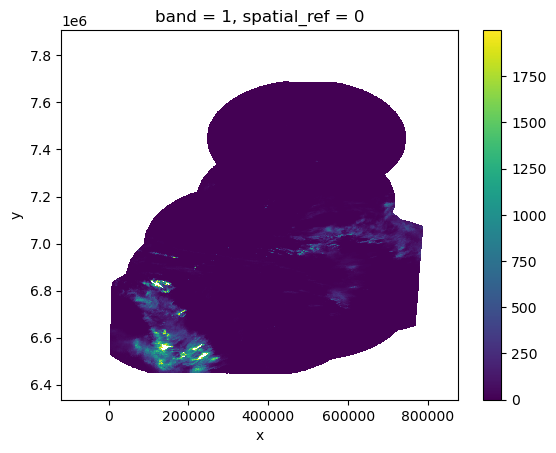
xds = rioxarray.open_rasterio(fs.open(url))
xds
<xarray.DataArray (band: 1, y: 1345, x: 850)> Size: 2MB
[1143250 values with dtype=uint16]
Coordinates:
* band (band) int64 8B 1
* x (x) float64 7kB -1.177e+05 -1.166e+05 ... 8.738e+05 8.75e+05
* y (y) float64 11kB 7.907e+06 7.906e+06 ... 6.337e+06 6.336e+06
spatial_ref int64 8B 0
Attributes:
GDAL_METADATA: <GDALMetadata>\n<Item name="Observation time" format="YYY...
AREA_OR_POINT: Area
scale_factor: 1.0
add_offset: 0.0xds.isel(band=0).where(xds < 2000).plot()
<matplotlib.collections.QuadMesh at 0x7fb938c334c0>
Create Input File List
Here we are using fsspec's glob functionality along with the * wildcard operator and some string slicing to grab a list of GeoTIFF files from a s3 fsspec filesystem.
# Initiate fsspec filesystems for reading
fs_read = fsspec.filesystem("s3", anon=True, skip_instance_cache=True)
files_paths = fs_read.glob(
"s3://fmi-opendata-radar-geotiff/2023/01/01/FIN-ACRR-3067-1KM/*24H-3067-1KM.tif"
)
# Here we prepend the prefix 's3://', which points to AWS.
files_paths = sorted(["s3://" + f for f in files_paths])
Start a Dask Client
To parallelize the creation of our reference files, we will use Dask. For a detailed guide on how to use Dask and Kerchunk, see the Foundations notebook: Kerchunk and Dask.
client = Client(n_workers=8, silence_logs=logging.ERROR)
client
Client
Client-d68bb06f-b0ee-11ef-8cdf-7c1e5222ecf8
| Connection method: Cluster object | Cluster type: distributed.LocalCluster |
| Dashboard: http://127.0.0.1:8787/status |
Cluster Info
LocalCluster
529afdab
| Dashboard: http://127.0.0.1:8787/status | Workers: 8 |
| Total threads: 8 | Total memory: 15.61 GiB |
| Status: running | Using processes: True |
Scheduler Info
Scheduler
Scheduler-38004c7e-36af-4c3a-90fa-8cdb7356d0ca
| Comm: tcp://127.0.0.1:43443 | Workers: 8 |
| Dashboard: http://127.0.0.1:8787/status | Total threads: 8 |
| Started: Just now | Total memory: 15.61 GiB |
Workers
Worker: 0
| Comm: tcp://127.0.0.1:38243 | Total threads: 1 |
| Dashboard: http://127.0.0.1:40241/status | Memory: 1.95 GiB |
| Nanny: tcp://127.0.0.1:45345 | |
| Local directory: /tmp/dask-scratch-space/worker-0tnublw0 | |
Worker: 1
| Comm: tcp://127.0.0.1:36741 | Total threads: 1 |
| Dashboard: http://127.0.0.1:37629/status | Memory: 1.95 GiB |
| Nanny: tcp://127.0.0.1:34977 | |
| Local directory: /tmp/dask-scratch-space/worker-ahz0gtbr | |
Worker: 2
| Comm: tcp://127.0.0.1:45587 | Total threads: 1 |
| Dashboard: http://127.0.0.1:34865/status | Memory: 1.95 GiB |
| Nanny: tcp://127.0.0.1:35621 | |
| Local directory: /tmp/dask-scratch-space/worker-imqszvku | |
Worker: 3
| Comm: tcp://127.0.0.1:43483 | Total threads: 1 |
| Dashboard: http://127.0.0.1:46287/status | Memory: 1.95 GiB |
| Nanny: tcp://127.0.0.1:34117 | |
| Local directory: /tmp/dask-scratch-space/worker-jqoo9bst | |
Worker: 4
| Comm: tcp://127.0.0.1:38623 | Total threads: 1 |
| Dashboard: http://127.0.0.1:34921/status | Memory: 1.95 GiB |
| Nanny: tcp://127.0.0.1:34439 | |
| Local directory: /tmp/dask-scratch-space/worker-9nn4gnve | |
Worker: 5
| Comm: tcp://127.0.0.1:33911 | Total threads: 1 |
| Dashboard: http://127.0.0.1:43329/status | Memory: 1.95 GiB |
| Nanny: tcp://127.0.0.1:40101 | |
| Local directory: /tmp/dask-scratch-space/worker-rim13nh2 | |
Worker: 6
| Comm: tcp://127.0.0.1:43991 | Total threads: 1 |
| Dashboard: http://127.0.0.1:41865/status | Memory: 1.95 GiB |
| Nanny: tcp://127.0.0.1:37671 | |
| Local directory: /tmp/dask-scratch-space/worker-ocy8wg1v | |
Worker: 7
| Comm: tcp://127.0.0.1:33143 | Total threads: 1 |
| Dashboard: http://127.0.0.1:33439/status | Memory: 1.95 GiB |
| Nanny: tcp://127.0.0.1:37755 | |
| Local directory: /tmp/dask-scratch-space/worker-_q1jxtoe | |
def generate_virtual_dataset(file):
storage_options = dict(
anon=True, default_fill_cache=False, default_cache_type="none"
)
vds = open_virtual_dataset(
file,
indexes={},
filetype="tiff",
reader_options={
"remote_options": {"anon": True},
"storage_options": storage_options,
},
)
# Pre-process virtual datasets to extract time step information from the filename
subst = file.split("/")[-1].split(".json")[0].split("_")[0]
time_val = datetime.strptime(subst, "%Y%m%d%H%M")
vds = vds.expand_dims(dim={"time": [time_val]})
# Only include the raw data, not the overviews
vds = vds[["0"]]
return vds
# Generate Dask Delayed objects
tasks = [dask.delayed(generate_virtual_dataset)(file) for file in files_paths]
# Start parallel processing
import warnings
warnings.filterwarnings("ignore")
virtual_datasets = dask.compute(*tasks)
/home/runner/miniconda3/envs/kerchunk-cookbook/lib/python3.10/site-packages/virtualizarr/readers/tiff.py:41: UserWarning: storage_options have been dropped from reader_options as they are not supported by kerchunk.tiff.tiff_to_zarr
warnings.warn(
/home/runner/miniconda3/envs/kerchunk-cookbook/lib/python3.10/site-packages/virtualizarr/readers/tiff.py:41: UserWarning: storage_options have been dropped from reader_options as they are not supported by kerchunk.tiff.tiff_to_zarr
warnings.warn(
/home/runner/miniconda3/envs/kerchunk-cookbook/lib/python3.10/site-packages/virtualizarr/readers/tiff.py:41: UserWarning: storage_options have been dropped from reader_options as they are not supported by kerchunk.tiff.tiff_to_zarr
warnings.warn(
/home/runner/miniconda3/envs/kerchunk-cookbook/lib/python3.10/site-packages/virtualizarr/readers/tiff.py:41: UserWarning: storage_options have been dropped from reader_options as they are not supported by kerchunk.tiff.tiff_to_zarr
warnings.warn(
/home/runner/miniconda3/envs/kerchunk-cookbook/lib/python3.10/site-packages/virtualizarr/readers/tiff.py:41: UserWarning: storage_options have been dropped from reader_options as they are not supported by kerchunk.tiff.tiff_to_zarr
warnings.warn(
/home/runner/miniconda3/envs/kerchunk-cookbook/lib/python3.10/site-packages/virtualizarr/readers/tiff.py:41: UserWarning: storage_options have been dropped from reader_options as they are not supported by kerchunk.tiff.tiff_to_zarr
warnings.warn(
/home/runner/miniconda3/envs/kerchunk-cookbook/lib/python3.10/site-packages/virtualizarr/readers/tiff.py:41: UserWarning: storage_options have been dropped from reader_options as they are not supported by kerchunk.tiff.tiff_to_zarr
warnings.warn(
/home/runner/miniconda3/envs/kerchunk-cookbook/lib/python3.10/site-packages/virtualizarr/readers/tiff.py:41: UserWarning: storage_options have been dropped from reader_options as they are not supported by kerchunk.tiff.tiff_to_zarr
warnings.warn(
/home/runner/miniconda3/envs/kerchunk-cookbook/lib/python3.10/site-packages/virtualizarr/readers/tiff.py:41: UserWarning: storage_options have been dropped from reader_options as they are not supported by kerchunk.tiff.tiff_to_zarr
warnings.warn(
/home/runner/miniconda3/envs/kerchunk-cookbook/lib/python3.10/site-packages/virtualizarr/readers/tiff.py:41: UserWarning: storage_options have been dropped from reader_options as they are not supported by kerchunk.tiff.tiff_to_zarr
warnings.warn(
/home/runner/miniconda3/envs/kerchunk-cookbook/lib/python3.10/site-packages/virtualizarr/readers/tiff.py:41: UserWarning: storage_options have been dropped from reader_options as they are not supported by kerchunk.tiff.tiff_to_zarr
warnings.warn(
/home/runner/miniconda3/envs/kerchunk-cookbook/lib/python3.10/site-packages/virtualizarr/readers/tiff.py:41: UserWarning: storage_options have been dropped from reader_options as they are not supported by kerchunk.tiff.tiff_to_zarr
warnings.warn(
/home/runner/miniconda3/envs/kerchunk-cookbook/lib/python3.10/site-packages/virtualizarr/readers/tiff.py:41: UserWarning: storage_options have been dropped from reader_options as they are not supported by kerchunk.tiff.tiff_to_zarr
warnings.warn(
/home/runner/miniconda3/envs/kerchunk-cookbook/lib/python3.10/site-packages/virtualizarr/readers/tiff.py:41: UserWarning: storage_options have been dropped from reader_options as they are not supported by kerchunk.tiff.tiff_to_zarr
warnings.warn(
/home/runner/miniconda3/envs/kerchunk-cookbook/lib/python3.10/site-packages/virtualizarr/readers/tiff.py:41: UserWarning: storage_options have been dropped from reader_options as they are not supported by kerchunk.tiff.tiff_to_zarr
warnings.warn(
/home/runner/miniconda3/envs/kerchunk-cookbook/lib/python3.10/site-packages/virtualizarr/readers/tiff.py:41: UserWarning: storage_options have been dropped from reader_options as they are not supported by kerchunk.tiff.tiff_to_zarr
warnings.warn(
/home/runner/miniconda3/envs/kerchunk-cookbook/lib/python3.10/site-packages/virtualizarr/readers/tiff.py:41: UserWarning: storage_options have been dropped from reader_options as they are not supported by kerchunk.tiff.tiff_to_zarr
warnings.warn(
/home/runner/miniconda3/envs/kerchunk-cookbook/lib/python3.10/site-packages/virtualizarr/readers/tiff.py:41: UserWarning: storage_options have been dropped from reader_options as they are not supported by kerchunk.tiff.tiff_to_zarr
warnings.warn(
/home/runner/miniconda3/envs/kerchunk-cookbook/lib/python3.10/site-packages/virtualizarr/readers/tiff.py:41: UserWarning: storage_options have been dropped from reader_options as they are not supported by kerchunk.tiff.tiff_to_zarr
warnings.warn(
/home/runner/miniconda3/envs/kerchunk-cookbook/lib/python3.10/site-packages/virtualizarr/readers/tiff.py:41: UserWarning: storage_options have been dropped from reader_options as they are not supported by kerchunk.tiff.tiff_to_zarr
warnings.warn(
/home/runner/miniconda3/envs/kerchunk-cookbook/lib/python3.10/site-packages/virtualizarr/readers/tiff.py:41: UserWarning: storage_options have been dropped from reader_options as they are not supported by kerchunk.tiff.tiff_to_zarr
warnings.warn(
/home/runner/miniconda3/envs/kerchunk-cookbook/lib/python3.10/site-packages/virtualizarr/readers/tiff.py:41: UserWarning: storage_options have been dropped from reader_options as they are not supported by kerchunk.tiff.tiff_to_zarr
warnings.warn(
/home/runner/miniconda3/envs/kerchunk-cookbook/lib/python3.10/site-packages/virtualizarr/readers/tiff.py:41: UserWarning: storage_options have been dropped from reader_options as they are not supported by kerchunk.tiff.tiff_to_zarr
warnings.warn(
Combine virtual datasets
combined_vds = xr.concat(virtual_datasets, dim="time")
combined_vds
<xarray.Dataset> Size: 53MB
Dimensions: (time: 23, Y: 1345, X: 850)
Coordinates:
* time (time) datetime64[ns] 184B 2023-01-01T01:00:00 ... 2023-01-01T23...
Dimensions without coordinates: Y, X
Data variables:
0 (time, Y, X) uint16 53MB ManifestArray<shape=(23, 1345, 850), dt...
Attributes: (12/15)
multiscales: [{'datasets': [{'path': '0'}, {'path': '1'}, {'p...
GDAL_METADATA: <GDALMetadata>\n<Item name="Observ...
KeyDirectoryVersion: 1
KeyRevision: 1
KeyRevisionMinor: 0
GTModelTypeGeoKey: 1
... ...
GeogAngularUnitsGeoKey: 9102
GeogTOWGS84GeoKey: [0.0, 0.0, 0.0]
ProjectedCSTypeGeoKey: 3067
ProjLinearUnitsGeoKey: 9001
ModelPixelScale: [1169.2930568410832, 1168.8701637541064, 0.0]
ModelTiepoint: [0.0, 0.0, 0.0, -118331.36640835612, 7907751.537...Shut down the Dask cluster
client.shutdown()