Source determination through NOAA HYSPLIT trajectory model
HYSPLIT models simulate the dispersion and trajectory of substances transported and dispersed through our atmosphere, over local to global scales.
A common application is a back trajectory analysis to determine the origin of air masses and establish source-receptor relationships.
# import libraries
import xarray as xr # for data storage and processing
import numpy as np # for data storage and processig
import pandas as pd # for data storage and processig
import act
from datetime import datetime # for formating date and time
import matplotlib.pyplot as plt
# define the function to read the HYSPLIT model output file
def read_hysplit(filename, base_year=2000):
"""
Reads an input HYSPLIT trajectory for plotting in ACT.
Parameters
----------
filename: str
The input file name.
base_year: int
The first year of the century in which the data are contained.
Returns
-------
ds: xarray Dataset
The ACT dataset containing the HYSPLIT trajectories
"""
ds = xr.Dataset({})
num_lines = 0
with open(filename) as filebuf:
num_grids = int(filebuf.readline().split()[0])
num_lines += 1
grid_times = []
grid_names = []
forecast_hours = np.zeros(num_grids)
for i in range(num_grids):
data = filebuf.readline().split()
num_lines += 1
grid_names.append(data[0])
grid_times.append(
datetime(
year=(int(data[1]) + base_year),
month=int(data[2]),
day=int(data[3]),
hour=int(data[4]),
)
)
forecast_hours[i] = int(data[5])
ds["grid_forecast_hour"] = xr.DataArray(forecast_hours, dims=["num_grids"])
ds["grid_forecast_hour"].attrs["standard_name"] = "Grid forecast hour"
ds["grid_forecast_hour"].attrs["units"] = "Hour [UTC]"
ds["grid_times"] = xr.DataArray(np.array(grid_times), dims=["num_grids"])
data_line = filebuf.readline().split()
num_lines += 1
ds.attrs["trajectory_direction"] = data_line[1]
ds.attrs["vertical_motion_calculation_method"] = data_line[2]
num_traj = int(data_line[0])
traj_times = []
start_lats = np.zeros(num_traj)
start_lons = np.zeros(num_traj)
start_alt = np.zeros(num_traj)
for i in range(num_traj):
data = filebuf.readline().split()
num_lines += 1
traj_times.append(
datetime(
year=(base_year + int(data[0])),
month=int(data[1]),
day=int(data[2]),
hour=int(data[3]),
)
)
start_lats[i] = float(data[4])
start_lons[i] = float(data[5])
start_alt[i] = float(data[6])
ds["start_latitude"] = xr.DataArray(start_lats, dims=["num_trajectories"])
ds["start_latitude"].attrs["long_name"] = "Trajectory start latitude"
ds["start_latitude"].attrs["units"] = "degree"
ds["start_longitude"] = xr.DataArray(start_lats, dims=["num_trajectories"])
ds["start_longitude"].attrs["long_name"] = "Trajectory start longitude"
ds["start_longitude"].attrs["units"] = "degree"
ds["start_altitude"] = xr.DataArray(start_alt, dims=["num_trajectories"])
ds["start_altitude"].attrs["long_name"] = "Trajectory start altitude"
ds["start_altitude"].attrs["units"] = "degree"
data = filebuf.readline().split()
num_lines += 1
var_list = [
"trajectory_number",
"grid_number",
"year",
"month",
"day",
"hour",
"minute",
"forecast_hour",
"age",
"lat",
"lon",
"alt",
]
for variable in data[1:]:
var_list.append(variable)
input_df = pd.read_csv(
filebuf, sep=r'\s+', index_col=False, names=var_list, skiprows=1
) # noqa W605
input_df['year'] = base_year + input_df['year']
input_df['year'] = input_df['year'].astype(int)
input_df['month'] = input_df['month'].astype(int)
input_df['day'] = input_df['day'].astype(int)
input_df['hour'] = input_df['hour'].astype(int)
input_df['minute'] = input_df['minute'].astype(int)
input_df['time'] = pd.to_datetime(
input_df[["year", "month", "day", "hour", "minute"]], format='%y%m%d%H%M'
)
input_df = input_df.set_index("time")
del input_df["year"]
del input_df["month"]
del input_df["day"]
del input_df["hour"]
del input_df["minute"]
ds = ds.merge(input_df.to_xarray())
ds.attrs['datastream'] = 'hysplit'
ds["trajectory_number"].attrs["standard_name"] = "Trajectory number"
ds["trajectory_number"].attrs["units"] = "1"
ds["grid_number"].attrs["standard_name"] = "Grid number"
ds["grid_number"].attrs["units"] = "1"
ds["age"].attrs["standard_name"] = "Grid number"
ds["age"].attrs["units"] = "1"
ds["lat"].attrs["standard_name"] = "Latitude"
ds["lat"].attrs["units"] = "degree"
ds["lon"].attrs["standard_name"] = "Longitude"
ds["lon"].attrs["units"] = "degree"
ds["alt"].attrs["standard_name"] = "Altitude"
ds["alt"].attrs["units"] = "meter"
return ds
Plotting HYSPLIT back trajectory and compare with NASA firemap
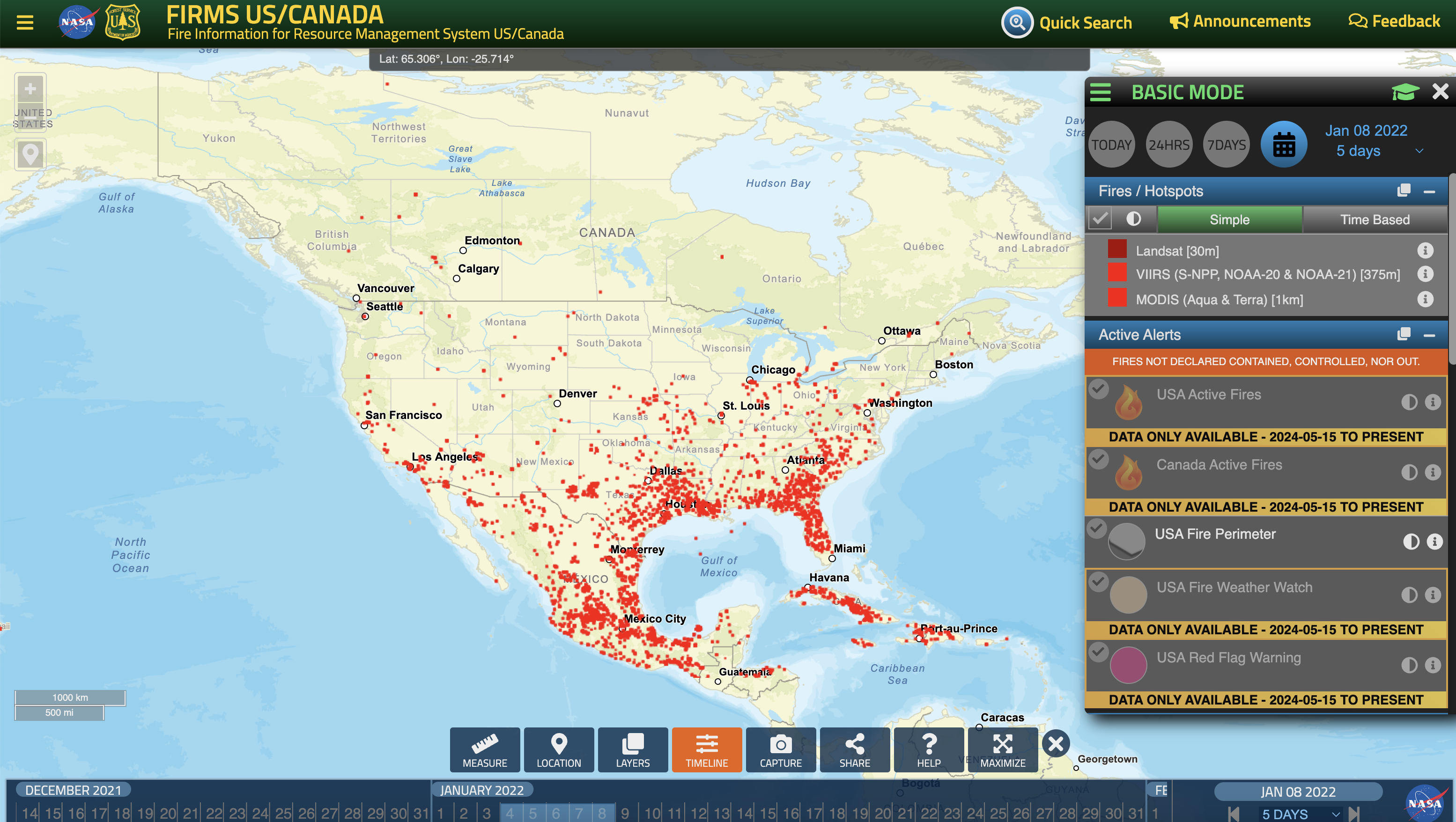
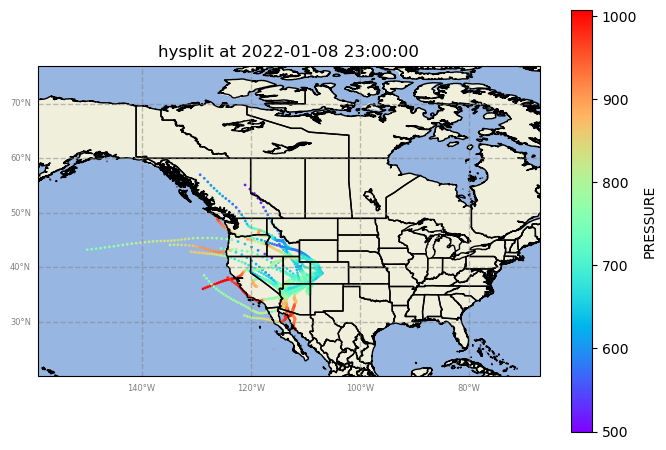
Case 1: Jan 8th, 2022
# Load the data
filename = 'guc_010822.txt'
ds = read_hysplit(filename)
# Use the GeographicPlotDisplay object to make the plot
disp = act.plotting.GeographicPlotDisplay(ds)
ax = disp.geoplot('PRESSURE', cartopy_feature=['STATES', 'OCEAN', 'LAND'], s = 1)
ax.set_xlim([-159, -67])
ax.set_ylim([20, 77])
plt.show()
Fire Map Link
![]()
Click here to view the interactive map
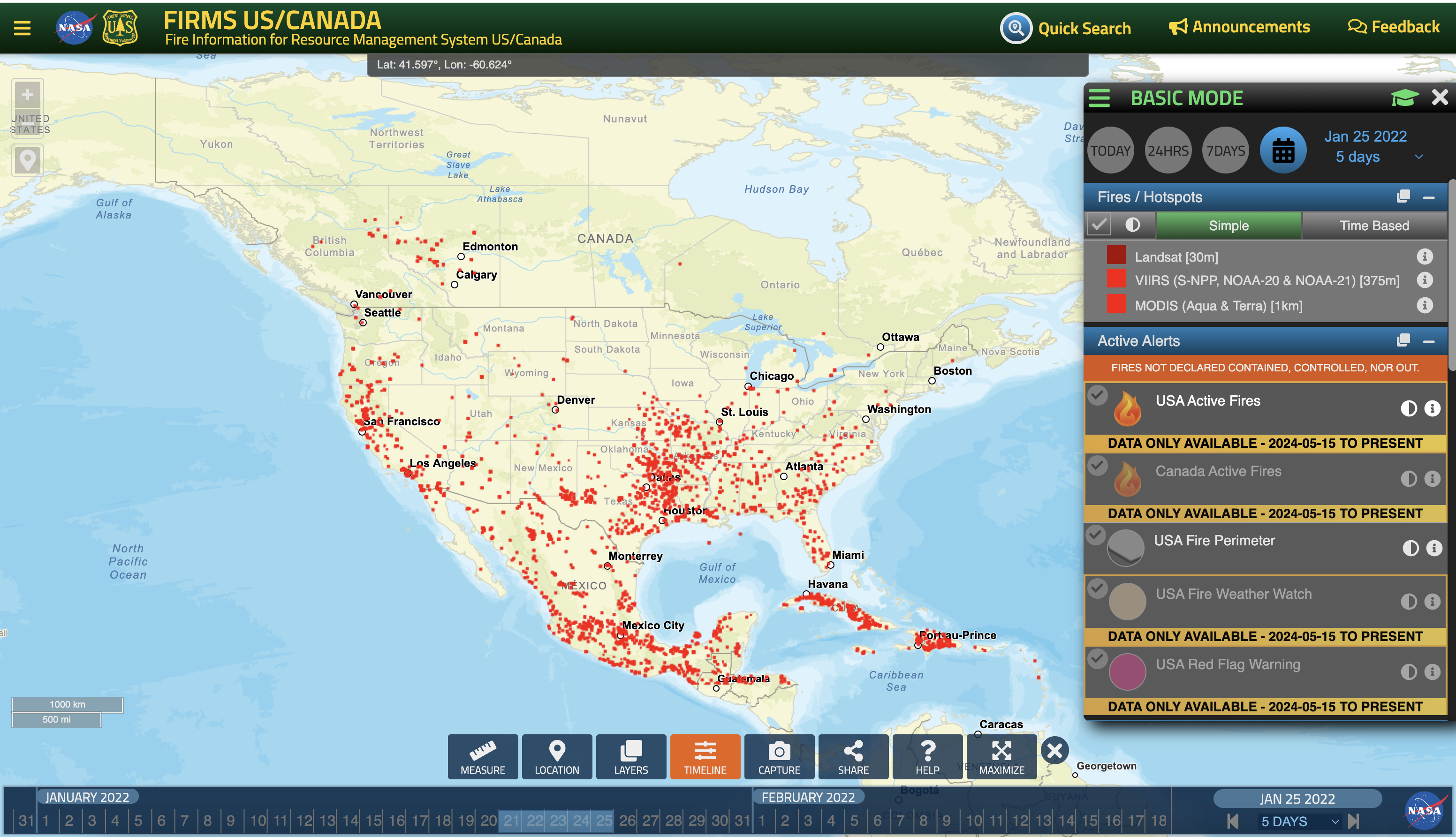
Case 2: Jan 25th, 2022
# Load the data
filename = 'guc_012522.txt'
ds = read_hysplit(filename)
# Use the GeographicPlotDisplay object to make the plot
disp = act.plotting.GeographicPlotDisplay(ds)
ax = disp.geoplot('PRESSURE', cartopy_feature=['STATES', 'OCEAN', 'LAND'], s = 1)
ax.set_xlim([-159, -67])
ax.set_ylim([20, 77])
plt.show()
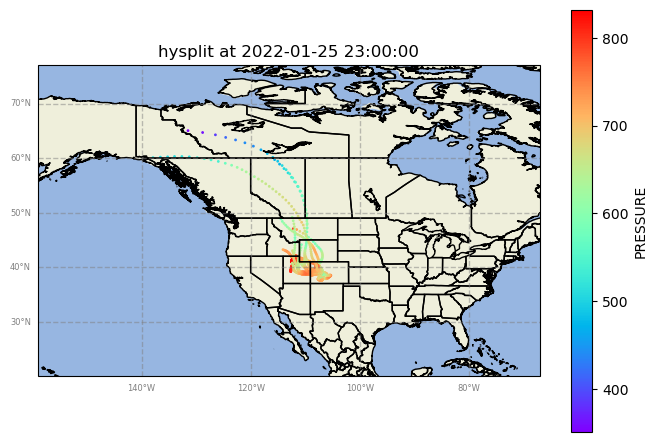
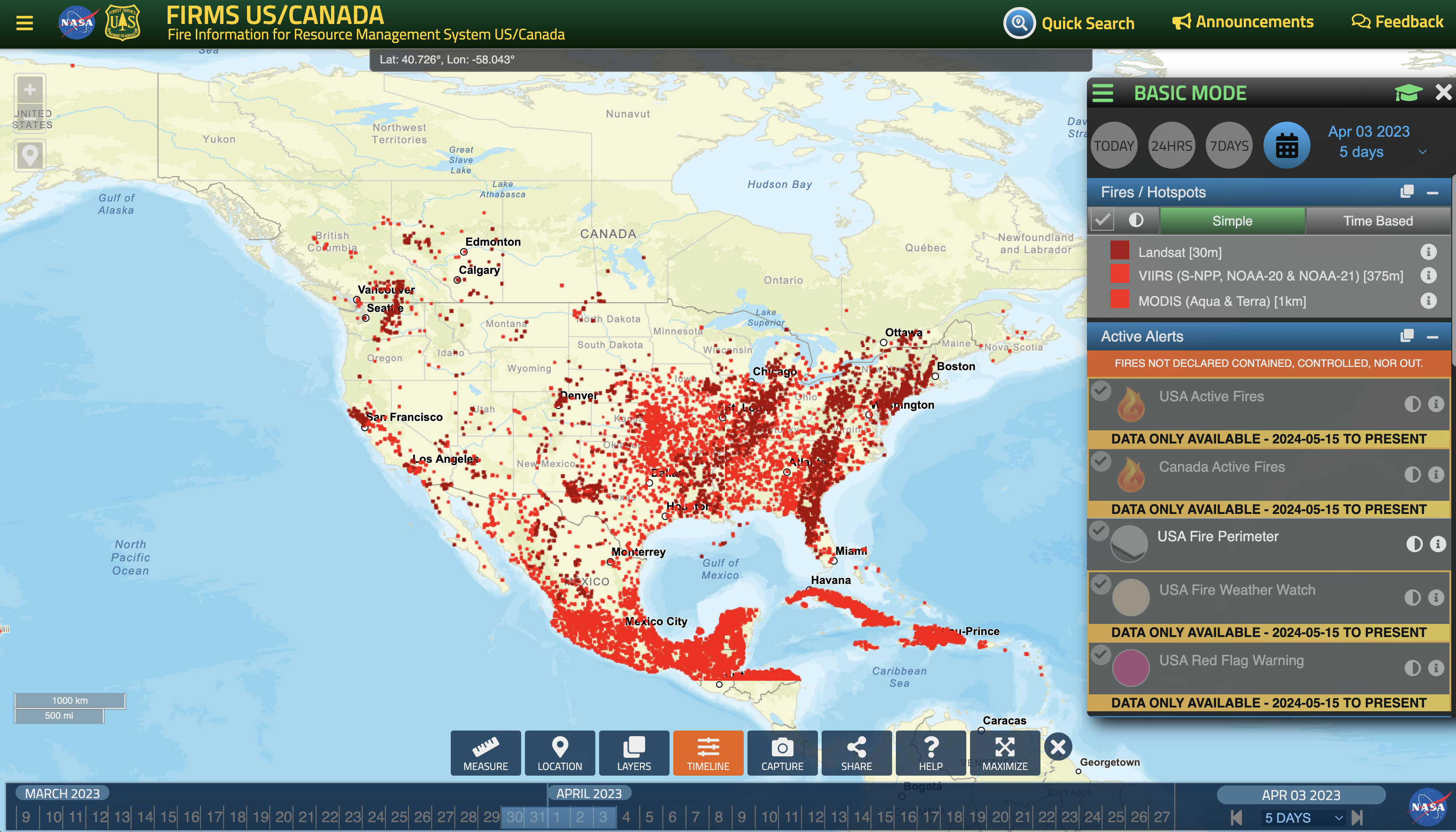
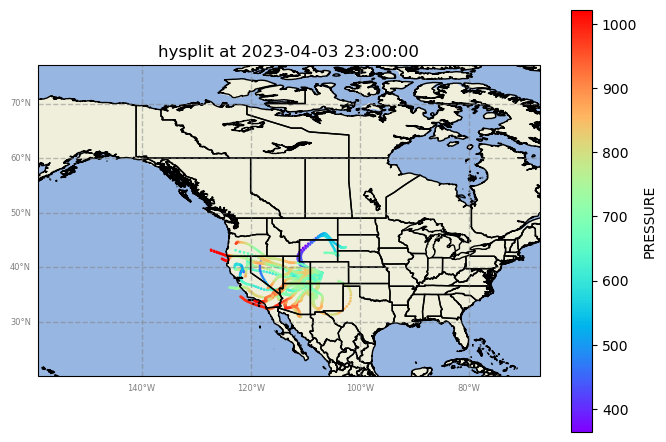
Case 3: Apr 3rd, 2023
# Load the data
filename = 'guc_040323.txt'
ds = read_hysplit(filename)
# Use the GeographicPlotDisplay object to make the plot
disp = act.plotting.GeographicPlotDisplay(ds)
ax = disp.geoplot('PRESSURE', cartopy_feature=['STATES', 'OCEAN', 'LAND'], s = 1)
ax.set_xlim([-159, -67])
ax.set_ylim([20, 77])
plt.show()
from IPython.display import display, HTML
# Adjust the path to your HTML file accordingly
file_path = "ARM_smps_data.html"
# Read the content of the HTML file
with open(file_path, 'r') as file:
html_content = file.read()
# Display the HTML content
display(HTML(html_content))
ARM_SAIL_project
Joy Lai
2024-05-23
load libraries
library(ncdf4) # package for netcdf manipulation
library(ggplot2) # package for plotting
library(data.table) #read data into datatble or creat datatble
library(patchwork) # arrange plot_layout
library(janitor) #clean data table names##
## Attaching package: 'janitor'## The following objects are masked from 'package:stats':
##
## chisq.test, fisher.testlibrary(tidyr) # to use function pivot_longer
library(dplyr) ## to use function group_by ##
## Attaching package: 'dplyr'## The following objects are masked from 'package:data.table':
##
## between, first, last## The following objects are masked from 'package:stats':
##
## filter, lag## The following objects are masked from 'package:base':
##
## intersect, setdiff, setequal, unionlibrary(viridis)## Loading required package: viridisLite##E Batch mode processing all .nc files
# Define a function to read .nc files and extract cpc variables
smps_read_nc_file <- function(file_path) {
#S4_alt <- 2764 # the height of S4 site above sea level
nc_data <- nc_open(file_path)
smps.totalconc <- ncvar_get(nc_data, "total_N_conc")
smps.alt <- ncvar_get(nc_data, "alt")
date_data <- ncvar_get(nc_data, "base_time")
time_data <- ncvar_get(nc_data, "time_offset")
# Convert time data to POSIXct (datetime) objects
smps.datetime <- as.POSIXct(time_data, origin = date_data, tz = "UTC")
nc_close(nc_data)
return(list(smps.datetime = smps.datetime, smps.totalconc = smps.totalconc, smps.alt = smps.alt))
}
# Define a function to create the plot
smps_create_plot <- function(smps.datetime, smps.totalconc, smps.alt, file_name) {
df <- data.frame(smps.datetime = smps.datetime, smps.totalconc = smps.totalconc, smps.alt = smps.alt)
ggplot(df, aes(x = smps.datetime, y = smps.totalconc)) +
geom_line(size = 2)+
scale_y_log10()+
labs(x = "Datetime", y = "SMPS total num conc (#/cc)", title = paste("SMPS Total Concentration vs. Date -", file_name)) +
theme_minimal()
}filepath <- "/Users/laiz097/Library/CloudStorage/OneDrive-PNNL/Documents/2024/ARM_summer_school_2024/SAIL_group_project/SMPS_data"
file_list <- list.files(path = filepath, pattern = ".nc", full.names=TRUE)
# Read each .nc file
nc_data_list <- lapply(file_list, nc_open)
# Loop through each file, read data, and create plots
for (file in file_list) {
data <- smps_read_nc_file(file)
plot <- smps_create_plot(data$smps.datetime, data$smps.totalconc, data$smps.alt, basename(file))
print(plot)
}## Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
## ℹ Please use `linewidth` instead.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
## generated.